| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:25:59 UTC |
|---|
| Update Date | 2020-05-21 16:28:41 UTC |
|---|
| BMDB ID | BMDB0006790 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Galactosylglycerol |
|---|
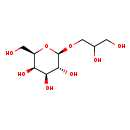
| Description | Galactosylglycerol, also known as galactosylglycerol, belongs to the class of organic compounds known as glycosylglycerols. These are glycerolipids structurally characterized by the presence of one or more sugar residues attached to glycerol via a glycosidic linkage. Galactosylglycerol is possibly soluble (in water) and an extremely weak basic (essentially neutral) compound (based on its pKa). Galactosylglycerol exists in all living species, ranging from bacteria to humans. Galactosylglycerol can be biosynthesized from D-galactose and glycerol; which is catalyzed by the enzyme Alpha-galactosidase a. In cattle, galactosylglycerol is involved in the metabolic pathway called the galactose metabolism pathway. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2R,3R,4S,5R,6R)-2-[(2R)-2,3-Dihydroxypropoxy]-6-(hydroxymethyl)oxane-3,4,5-triol | HMDB | | 3-b-D-Galactosyl-sn-glycerol | HMDB | | 3-beta-D-Galactosyl-sn-glycerol | HMDB | | 3-beta-delta-Galactosyl-sn-glycerol | HMDB | | Azepan-2-one | HMDB |
|
|---|
| Chemical Formula | C9H18O8 |
|---|
| Average Molecular Weight | 254.2344 |
|---|
| Monoisotopic Molecular Weight | 254.100167552 |
|---|
| IUPAC Name | (2R,3R,4S,5R,6R)-2-(2,3-dihydroxypropoxy)-6-(hydroxymethyl)oxane-3,4,5-triol |
|---|
| Traditional Name | galactosylglycerol |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | OCC(O)CO[C@@H]1O[C@H](CO)[C@H](O)[C@H](O)[C@H]1O |
|---|
| InChI Identifier | InChI=1S/C9H18O8/c10-1-4(12)3-16-9-8(15)7(14)6(13)5(2-11)17-9/h4-15H,1-3H2/t4?,5-,6+,7+,8-,9-/m1/s1 |
|---|
| InChI Key | NHJUPBDCSOGIKX-VGPGGAHRSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glycosylglycerols. These are glycerolipids structurally characterized by the presence of one or more sugar residues attached to glycerol via a glycosidic linkage. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Glycerolipids |
|---|
| Sub Class | Glycosylglycerols |
|---|
| Direct Parent | Glycosylglycerols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glycosylglycerol
- Glycosyl compound
- O-glycosyl compound
- Monosaccharide
- Oxane
- Secondary alcohol
- Polyol
- Oxacycle
- Acetal
- Organoheterocyclic compound
- Primary alcohol
- Alcohol
- Organooxygen compound
- Hydrocarbon derivative
- Organic oxygen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01pc-8950000000-2ab0ad026160a115d34a | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (6 TMS) - 70eV, Positive | splash10-00di-1120229000-fb00db4830a4b4ea861f | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4r-2290000000-2a5149feb24795c7b79d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004l-9230000000-2a96dc687f7a244ae29a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-004l-9500000000-ec5346c07e7023876c0d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-5690000000-6778794a074e42e6e8fd | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0itc-7930000000-60a6b3e596cefbf15555 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-006x-9300000000-67ad70b9f13e121d5e0c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0w29-1690000000-8198e0d9b54e0889ea5c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0r0c-9430000000-12dd3c060d7e8ad406ef | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-393bf60f5c57a1c20727 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-052r-0390000000-eeb61f13ce6373429218 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-9530000000-caee0578db41f8d047b0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-056r-9000000000-66eeaad1c45d16391f48 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|