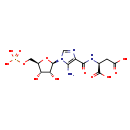| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:36:44 UTC |
|---|
| Update Date | 2020-05-21 16:28:53 UTC |
|---|
| BMDB ID | BMDB0000797 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | SAICAR |
|---|
| Description | SAICAR, also known as saicaribotide or succino-aicar, belongs to the class of organic compounds known as 1-ribosyl-imidazolecarboxamides. These are organic compounds containing the imidazole ring linked to a ribose ring through a 1-2 bond. SAICAR is a strong basic compound (based on its pKa). SAICAR exists in all eukaryotes, ranging from yeast to humans. SAICAR is a potentially toxic compound. SAICAR, with regard to humans, has been linked to several inborn metabolic disorders including fumarase deficiency and atic deficiency. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S)-2-[5-Amino-1-(5-phospho-beta-D-ribosyl)imidazole-4-carboxamido]succinic acid | ChEBI | | (S)-2-[5-Amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamido]succinate | ChEBI | | 1-(5'-Phosphoribosyl)-4-(N-succinocarboxamide)-5-aminoimidazole | ChEBI | | 1-(5'-Phosphoribosyl)-5-amino-4-(N-succinocarboxamide)-imidazole | ChEBI | | 5'-Phosphoribosyl-4-(N-succinocarboxamide)-5-aminoimidazole | ChEBI | | Succino-aicar | ChEBI | | Succinyl-5-aminoimidazole-4-carboxamide-1-ribose-5-phosphate | ChEBI | | Succinylaminoimidazolecarboxamide ribose-5'-phosphate | ChEBI | | (2S)-2-[5-Amino-1-(5-phospho-b-D-ribosyl)imidazole-4-carboxamido]succinate | Generator | | (2S)-2-[5-Amino-1-(5-phospho-b-D-ribosyl)imidazole-4-carboxamido]succinic acid | Generator | | (2S)-2-[5-Amino-1-(5-phospho-beta-D-ribosyl)imidazole-4-carboxamido]succinate | Generator | | (2S)-2-[5-Amino-1-(5-phospho-β-D-ribosyl)imidazole-4-carboxamido]succinate | Generator | | (2S)-2-[5-Amino-1-(5-phospho-β-D-ribosyl)imidazole-4-carboxamido]succinic acid | Generator | | (S)-2-[5-Amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamido]succinic acid | Generator | | Succinyl-5-aminoimidazole-4-carboxamide-1-ribose-5-phosphoric acid | Generator | | Succinylaminoimidazolecarboxamide ribose-5'-phosphoric acid | Generator | | (S)-2-(5-Amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamido)succinate | HMDB | | (S)-2-(5-Amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxamido)succinic acid | HMDB | | (S)-2-(5-Amino-1-(5-phospho-delta-ribosyl)imidazole-4-carboxamido)succinate | HMDB | | (S)-2-(5-Amino-1-(5-phospho-delta-ribosyl)imidazole-4-carboxamido)succinic acid | HMDB | | (S)-2-[5-Amino-1-(5-phospho-delta-ribosyl)imidazole-4-carboxamido]succinate | HMDB | | 1-(5'-Phosphoribosyl)-5-amino-4-(N-succinocarboxamide)-imidazole' 1-(5'-phosphoribosyl)-4-(N-succinocarboxamide)-5-aminoimidazole | HMDB | | 5'-Phosphoribosyl-4-(N-succinocarbozamide)-5-aminoimidazole | HMDB | | 5-Amino-4-imidazole-N-succinocarboxamide ribonucleotide | HMDB | | L-N-[(5-Amino-1-b-D-ribofuranosylimidazol-4-yl)carbonyl]-5'-(dihydrogen phosphate) | HMDB | | L-N-[(5-Amino-1-beta-delta-ribofuranosylimidazol-4-yl)carbonyl]-5'-(dihydrogen phosphate) | HMDB | | N-(5-Amino-1-ribofuranosylimidazol-4-ylcarbonyl)aspartic acid 5'-phosphate | HMDB | | N-[5-Amino-1-(5'-phosphoribofuranosyl)-4-imidazolecarbonyl]aspartate | HMDB | | N-[5-Amino-1-(5'-phosphoribofuranosyl)-4-imidazolecarbonyl]aspartic acid | HMDB | | Phosphoribosylaminoimidazolesuccinocarboxamide | HMDB | | N-(5-Amino-1-beta-D-ribofuranosylimidazole-4-carbonyl)-L-aspartic acid 5'-phosphate | HMDB | | 5'-Phosphoribosyl-4-(N-succinylcarboxamide)-5-aminoimidazole | HMDB | | SAICAR, (D)-isomer | HMDB | | SAICAribotide | HMDB | | Succinylaminoimidazole carboxamide ribotide | HMDB | | SAICA ribotide | HMDB | | Succinylaminoimidazolecarboxamide ribose-5’-phosphate | HMDB |
|
|---|
| Chemical Formula | C13H19N4O12P |
|---|
| Average Molecular Weight | 454.2833 |
|---|
| Monoisotopic Molecular Weight | 454.073708604 |
|---|
| IUPAC Name | (2S)-2-({5-amino-1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-[(phosphonooxy)methyl]oxolan-2-yl]-1H-imidazol-4-yl}formamido)butanedioic acid |
|---|
| Traditional Name | saicar |
|---|
| CAS Registry Number | 3031-95-6 |
|---|
| SMILES | NC1=C(N=CN1[C@@H]1O[C@H](COP(O)(O)=O)[C@@H](O)[C@H]1O)C(=O)N[C@@H](CC(O)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C13H19N4O12P/c14-10-7(11(22)16-4(13(23)24)1-6(18)19)15-3-17(10)12-9(21)8(20)5(29-12)2-28-30(25,26)27/h3-5,8-9,12,20-21H,1-2,14H2,(H,16,22)(H,18,19)(H,23,24)(H2,25,26,27)/t4-,5+,8+,9+,12+/m0/s1 |
|---|
| InChI Key | NAQGHJTUZRHGAC-ZZZDFHIKSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 1-ribosyl-imidazolecarboxamides. These are organic compounds containing the imidazole ring linked to a ribose ring through a 1-2 bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Imidazole ribonucleosides and ribonucleotides |
|---|
| Sub Class | 1-ribosyl-imidazolecarboxamides |
|---|
| Direct Parent | 1-ribosyl-imidazolecarboxamides |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1-ribosyl-imidazolecarboxamide
- Pentose-5-phosphate
- Pentose phosphate
- Aspartic acid or derivatives
- N-acyl-alpha amino acid or derivatives
- N-acyl-alpha-amino acid
- Glycosyl compound
- N-glycosyl compound
- Monosaccharide phosphate
- Alpha-amino acid or derivatives
- Pentose monosaccharide
- 2-heteroaryl carboxamide
- Imidazolyl carboxylic acid derivative
- Imidazole-4-carbonyl group
- Monoalkyl phosphate
- N-substituted imidazole
- Organic phosphoric acid derivative
- Alkyl phosphate
- Dicarboxylic acid or derivatives
- Phosphoric acid ester
- Aminoimidazole
- Monosaccharide
- Imidazole
- Heteroaromatic compound
- Azole
- Vinylogous amide
- Tetrahydrofuran
- Amino acid or derivatives
- Carboxamide group
- Amino acid
- 1,2-diol
- Secondary carboxylic acid amide
- Secondary alcohol
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Carboxylic acid derivative
- Carboxylic acid
- Alcohol
- Organic oxygen compound
- Carbonyl group
- Organic nitrogen compound
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Amine
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-05n1-9732500000-f3a7f13b1dee5f81c7a8 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-05ot-9600116000-74a62efcb34072bbe352 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-055o-1592600000-05db249dfb0f14143f9f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-3950000000-6ebf6a64483dd1392782 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001l-5940000000-e218e278f5195eb90341 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0ug3-6381900000-4fec69ff813fe652ea7d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004j-9350000000-e4e387627fbca1ef1201 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9100000000-95f6153e96335a5d2b32 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0f79-2012900000-f38ec5261c12799bc354 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9022000000-a24939a2980b6834f5bc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9110000000-7f66eadbb3c3527bbc8c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0024900000-6f3c27fcd647c50d79d0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0096-1293000000-d46017274b02e79e4277 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-3891000000-ed7da10b4a914197adb3 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| Synthesis Reference | Shaw, Gordon; Thomas, Peter S.; Patey, Carole A. H.; Thomas, Susan E. Purines, pyrimidines and imidazoles. Part 50. Inhibition of adenylosuccinate AMP-lyase no. 4.3.2.2. by derivatives of N-(5-amino-1-b-D-ribofuranosylimidazole-4-carbonyl)-L-aspartic acid 5'-phosphate (SAICAR) and virazole 5'-phosphate. Journal of the Chemical Society, Perkin Transactions 1: Organic and Bio-Organic Chemistry (1972-1999) (1979), (6), 1415-24. |
|---|