Showing metabocard for DG(14:0/0:0/16:1n7) (BMDB0096275)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2020-03-25 15:30:34 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2020-04-22 18:57:06 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BMDB ID | BMDB0096275 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers | None | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
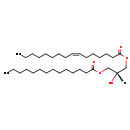
| Common Name | DG(14:0/0:0/16:1n7) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | DG(14:0/0:0/16:1n7) belongs to the family of Diacylglycerols. These are glycerolipids lipids containing a common glycerol backbone to which at least one fatty acyl group is esterified. It is involved in the phospholipid metabolic pathway. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C33H62O5 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight | 538.854 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight | 538.459725096 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | (2R)-2-hydroxy-3-(tetradecanoyloxy)propyl (7Z)-hexadec-7-enoate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | (2R)-2-hydroxy-3-(tetradecanoyloxy)propyl (7Z)-hexadec-7-enoate | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | [H][C@@](O)(COC(=O)CCCCCCCCCCCCC)COC(=O)CCCCC\C=C/CCCCCCCC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C33H62O5/c1-3-5-7-9-11-13-15-16-18-20-22-24-26-28-33(36)38-30-31(34)29-37-32(35)27-25-23-21-19-17-14-12-10-8-6-4-2/h16,18,31,34H,3-15,17,19-30H2,1-2H3/b18-16-/t31-/m1/s1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | IURSQZZYJNYILA-SYVZRPFGSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as 1,3-diacylglycerols. These are diacylglycerols containing a glycerol acylated at positions 1 and 3. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Lipids and lipid-like molecules | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Glycerolipids | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Diradylglycerols | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | 1,3-diacylglycerols | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Ontology | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Expected but not Quantified | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofunction | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Application | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biospecimen Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB0055960 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phenol Explorer Compound ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| FooDB ID | FDB098421 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KNApSAcK ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemspider ID | 74854338 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Compound ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BiGG ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| METLIN ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound | 131801693 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||