| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2020-03-10 17:02:58 UTC |
|---|
| Update Date | 2020-05-11 20:27:49 UTC |
|---|
| BMDB ID | BMDB0096208 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
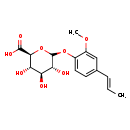
| Common Name | trans-isoeugenol-O-glucuronide |
|---|
| Description | trans-isoeugenol-O-glucuronide belongs to the class of organic compounds known as phenolic glycosides. These are organic compounds containing a phenolic structure attached to a glycosyl moiety. Some examples of phenolic structures include lignans, and flavonoids. Among the sugar units found in natural glycosides are D-glucose, L-Fructose, and L rhamnose. trans-isoeugenol-O-glucuronide is an extremely weak basic (essentially neutral) compound (based on its pKa). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S,3S,4S,5R)-3,4,5-Trihydroxy-6-{2-methoxy-4-[(1E)-prop-1-en-1-yl]phenoxy}oxane-2-carboxylate | Generator |
|
|---|
| Chemical Formula | C16H20O8 |
|---|
| Average Molecular Weight | 340.3252 |
|---|
| Monoisotopic Molecular Weight | 340.115817616 |
|---|
| IUPAC Name | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-{2-methoxy-4-[(1E)-prop-1-en-1-yl]phenoxy}oxane-2-carboxylic acid |
|---|
| Traditional Name | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-{2-methoxy-4-[(1E)-prop-1-en-1-yl]phenoxy}oxane-2-carboxylic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | COC1=CC(\C=C\C)=CC=C1OC1O[C@@H]([C@@H](O)[C@H](O)[C@H]1O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C16H20O8/c1-3-4-8-5-6-9(10(7-8)22-2)23-16-13(19)11(17)12(18)14(24-16)15(20)21/h3-7,11-14,16-19H,1-2H3,(H,20,21)/b4-3+/t11-,12-,13+,14-,16?/m0/s1 |
|---|
| InChI Key | HBQGHTRBYUFZLV-MIJQFFSFSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as phenolic glycosides. These are organic compounds containing a phenolic structure attached to a glycosyl moiety. Some examples of phenolic structures include lignans, and flavonoids. Among the sugar units found in natural glycosides are D-glucose, L-Fructose, and L rhamnose. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Phenolic glycosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Phenolic glycoside
- 1-o-glucuronide
- O-glucuronide
- Glucuronic acid or derivatives
- O-glycosyl compound
- Phenoxy compound
- Anisole
- Methoxybenzene
- Styrene
- Phenol ether
- Alkyl aryl ether
- Beta-hydroxy acid
- Monocyclic benzene moiety
- Hydroxy acid
- Monosaccharide
- Benzenoid
- Pyran
- Oxane
- Secondary alcohol
- Monocarboxylic acid or derivatives
- Organoheterocyclic compound
- Ether
- Carboxylic acid
- Carboxylic acid derivative
- Oxacycle
- Acetal
- Polyol
- Alcohol
- Carbonyl group
- Organic oxide
- Hydrocarbon derivative
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0abc-9263000000-28ef517776b4aebf3a15 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-03di-4111159000-9744a90b907a34da36d9 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01b9-0906000000-48cbd9daf96a67bbba35 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-0900000000-c3db8e6c9206bcc97a2c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00ke-4900000000-1717b89821326688d18d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01p9-1938000000-97bafd0181f2e3a2b850 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03dj-1911000000-ae2e1e0e7dcf9e248169 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-03ea-2900000000-61380e7246701c03fac0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0109000000-9e37c4f5a599e2fdb541 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01wf-0948000000-5a939ca8755a26e1893e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-017i-3900000000-d7f284fba3aed2db22c9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0009000000-0e256c0c28d005ee6300 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01ot-4924000000-a423a27ea1e11c166fb0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-056a-8941000000-c2f1b4cae7258f7da44f | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|