| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2020-03-10 17:02:11 UTC |
|---|
| Update Date | 2020-05-11 20:27:30 UTC |
|---|
| BMDB ID | BMDB0096160 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
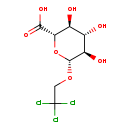
| Common Name | Trichloroethanol glucuronide |
|---|
| Description | Trichloroethanol glucuronide, also known as urochloralic acid or urochloralate, belongs to the class of organic compounds known as o-glucuronides. These are glucuronides in which the aglycone is linked to the carbohydrate unit through an O-glycosidic bond. Trichloroethanol glucuronide is an extremely weak basic (essentially neutral) compound (based on its pKa). These are compounds containing a glucuronic acid moeity (or a derivative), which consists of a glucose moiety with the C6 carbon oxidized to a carboxylic acid. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Urochloralic acid | Kegg | | 2,2,2-Trichloroethyl beta-D-glucopyranosiduronic acid | Kegg | | Urochloralate | Generator | | 2,2,2-Trichloroethyl b-D-glucopyranosiduronate | Generator | | 2,2,2-Trichloroethyl b-D-glucopyranosiduronic acid | Generator | | 2,2,2-Trichloroethyl beta-D-glucopyranosiduronate | Generator | | 2,2,2-Trichloroethyl β-D-glucopyranosiduronate | Generator | | 2,2,2-Trichloroethyl β-D-glucopyranosiduronic acid | Generator |
|
|---|
| Chemical Formula | C8H11Cl3O7 |
|---|
| Average Molecular Weight | 325.528 |
|---|
| Monoisotopic Molecular Weight | 323.957035827 |
|---|
| IUPAC Name | (2S,3S,4S,5R,6R)-3,4,5-trihydroxy-6-(2,2,2-trichloroethoxy)oxane-2-carboxylic acid |
|---|
| Traditional Name | trichloroethanol glucuronide |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | O[C@@H]1[C@@H](O)[C@H](OCC(Cl)(Cl)Cl)O[C@@H]([C@H]1O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C8H11Cl3O7/c9-8(10,11)1-17-7-4(14)2(12)3(13)5(18-7)6(15)16/h2-5,7,12-14H,1H2,(H,15,16)/t2-,3-,4+,5-,7+/m0/s1 |
|---|
| InChI Key | IQOASJJGUQMXDW-GHQVIJFQSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as o-glucuronides. These are glucuronides in which the aglycone is linked to the carbohydrate unit through an O-glycosidic bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | O-glucuronides |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1-o-glucuronide
- O-glucuronide
- Glycosyl compound
- O-glycosyl compound
- Beta-hydroxy acid
- Hydroxy acid
- Monosaccharide
- Pyran
- Oxane
- Secondary alcohol
- Acetal
- Carboxylic acid derivative
- Carboxylic acid
- Oxacycle
- Monocarboxylic acid or derivatives
- Organoheterocyclic compound
- Polyol
- Alcohol
- Hydrocarbon derivative
- Alkyl chloride
- Alkyl halide
- Organic oxide
- Carbonyl group
- Organochloride
- Organohalogen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4i-9211000000-495e7dd44cb7e4a28c2d | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-0005-7393461000-9058c49dc6f48c1446b1 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-05fr-0219000000-a4daa8fb6494256a6da5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-0911000000-7c87106fcc44781608f3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-06re-7910000000-b9a22f45fc1a6af28846 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-2749000000-1ebbf007253470bfd68b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00ba-5953000000-921524b5f29c7683f301 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0007-9700000000-904e7e9e2ebe8bdc7758 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00di-0009000000-55fab51f1680142a5e2b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ab9-0329000000-6e4ee7408afee8367eab | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03k9-9500000000-096906ff33fd7951adb0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0fk9-0009000000-b2836d5350aabaa31777 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05g0-7396000000-230a8ebdd8185b77fab2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9210000000-f503ef096bac83eedd1d | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|