| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2020-03-03 19:23:48 UTC |
|---|
| Update Date | 2020-04-22 15:55:15 UTC |
|---|
| BMDB ID | BMDB0063808 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Asparaginyl-Gamma-glutamate |
|---|
| Description | Asparaginyl-Gamma-glutamate, also known as N-ge dipeptide or asparaginyl-g-glutamic acid, belongs to the class of organic compounds known as glutamine and derivatives. Glutamine and derivatives are compounds containing glutamine or a derivative thereof resulting from reaction of glutamine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. Based on a literature review very few articles have been published on Asparaginyl-Gamma-glutamate. |
|---|
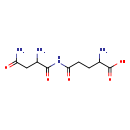
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Asparaginyl-g-glutamate | Generator | | Asparaginyl-g-glutamic acid | Generator | | Asparaginyl-gamma-glutamic acid | Generator | | Asparaginyl-γ-glutamate | Generator | | Asparaginyl-γ-glutamic acid | Generator | | Asn-gglu | HMDB | | Asparagine gamma-glutamate dipeptide | HMDB | | Asparagine-gamma-glutamate dipeptide | HMDB | | Asparaginylgamma-glutamate | HMDB | | L-Asparaginyl-L-gamma-glutamate | HMDB | | N-GE dipeptide | HMDB | | NGE dipeptide | HMDB | | 2-Amino-4-{[2-amino-3-(C-hydroxycarbonimidoyl)propanoyl]-C-hydroxycarbonimidoyl}butanoate | HMDB |
|
|---|
| Chemical Formula | C9H16N4O5 |
|---|
| Average Molecular Weight | 260.2471 |
|---|
| Monoisotopic Molecular Weight | 260.112069642 |
|---|
| IUPAC Name | 2-amino-4-[(2-amino-3-carbamoylpropanoyl)carbamoyl]butanoic acid |
|---|
| Traditional Name | 2-amino-4-[(2-amino-3-carbamoylpropanoyl)carbamoyl]butanoic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | NC(CCC(=O)NC(=O)C(N)CC(N)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C9H16N4O5/c10-4(9(17)18)1-2-7(15)13-8(16)5(11)3-6(12)14/h4-5H,1-3,10-11H2,(H2,12,14)(H,17,18)(H,13,15,16) |
|---|
| InChI Key | CXTYHSFBRYPJED-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glutamine and derivatives. Glutamine and derivatives are compounds containing glutamine or a derivative thereof resulting from reaction of glutamine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Glutamine and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glutamine or derivatives
- Asparagine or derivatives
- Alpha-amino acid amide
- Alpha-amino acid
- Fatty acyl
- Fatty acid
- Fatty amide
- N-acyl-amine
- Carboxylic acid imide
- Dicarboximide
- Carboxylic acid imide, n-unsubstituted
- Carboxamide group
- Amino acid
- Primary carboxylic acid amide
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic nitrogen compound
- Primary aliphatic amine
- Primary amine
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Carbonyl group
- Amine
- Organic oxygen compound
- Organooxygen compound
- Organonitrogen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-000i-9220000000-c3102c88122c1e1ecae0 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-014r-9260000000-9f307b7534c35a3af88f | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-015c-3790000000-8f6cf762ce572a67673e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fz1-7920000000-0ef9cc6ab940c7455309 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00dv-9100000000-6fd9d6bfcadd85b7692f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4l-0290000000-164d049a3acf14669fc7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-6950000000-b90598026c91677d2ba2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9400000000-7c352ff54b2e345702b8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0490000000-0e9380e36a5a062fd338 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0159-2960000000-d71231fab1065c58bb23 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0api-9600000000-b812c1956ab3954c59dc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4l-0390000000-7f0e2a5e22449352c846 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0037-5920000000-417f3cb9110867ea3c42 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9200000000-feba5f10ab702f8c1839 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|