| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2018-10-03 16:32:44 UTC |
|---|
| Update Date | 2020-06-04 20:40:38 UTC |
|---|
| BMDB ID | BMDB0063604 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | atrazine |
|---|
| Description | Atrazine is an organic compound consisting of an s-triazine-ring is a widely used herbicide. Its use is controversial due to widespread contamination in drinking water and its associations with birth defects and menstrual problems when consumed by humans at concentrations below government standards. Although it has been banned in the European Union,[2] it is still one of the most widely used herbicides in the world (Wikipedia). Atrazine is a suspected teratogen, causing demasculinization in male northern leopard frog even at low concentrations, and an estrogen disruptor. A 2010 study found that atrazine rendered 75 percent of male frogs sterile and turned one in 10 into females. A 2002 study found that exposure to atrazine caused male tadpoles to turn into hermaphrodites - frogs with both male and female sexual characteristics. But another study, requested by EPA and funded by Syngenta, was unable to reproduce these results. Atrazine was banned in the European Union (EU) in 2004 because of its persistent groundwater contamination. In the United States, however, atrazine is one of the most widely used herbicides, with 76 million pounds of it applied each year, in spite of the restriction that used to be imposed. Its endocrine disruptor effects, possible carcinogenic effect, and epidemiological connection to low sperm levels in men has led several researchers to call for banning it in the US.Rates of biodegradation are affected by atrazine's low solubility, thus surfactants may increase the degradation rate. Though the two alkyl moieties readily support growth of certain microorganisms, the atrazine ring is a poor energy source due to the oxidized state of ring carbon. In fact, the most common pathway for atrazine degradation involves the intermediate, cyanuric acid, in which carbon is fully oxidized, thus the ring is primarily a nitrogen source for aerobic microorganisms. Atrazine may be catabolized as a carbon and nitrogen source in reducing environments, and some aerobic atrazine degraders have been shown to use the compound for growth under anoxia in the presence of nitrate as an electron acceptor, a process referred to as a denitrification. When atrazine is used as a nitrogen source for bacterial growth, degradation may be regulated by the presence of alternative sources of nitrogen. In pure cultures of atrazine-degrading bacteria, as well as active soil communitites, atrazine ring nitrogen, but not carbon are assimilated into microbial biomass. Low concentrations of glucose can decrease the bioavailability, whereas higher concentrations promote the catabolism of atrazine. Tyrone Hayes, Department of Integrative Biology, University of California, notes that all of the studies that failed to conclude that atrazine caused hermaphroditism were plagued by poor experimental controls and were funded by Syngenta, one of the companies that produce the chemical. The U.S. Environmental Protection Agency (EPA) and its independent Scientific Advisory Panel (SAP) examined all available studies on this topic including Hayes' work and concluded that there are 'currently insufficient data' to determine if atrazine affects amphibian development. Hayes, formerly part of the SAP panel, resigned in 2000 to continue studies independently. The EPA and its SAP made recommendations concerning proper study design needed for further investigation into this issue. As required by the EPA, Syngenta conducted two experiments under Good Laboratory Practices (GLP) and inspection by the EPA and German regulatory authorities. The paper concluded 'These studies demonstrate that long-term exposure of larval X. laevis to atrazine at concentrations ranging from 0.01 to 100 microg/l does not affect growth, larval development, or sexual differentiation.' Another independent study in 2008 determined that 'the failure of recent studies to find that atrazine feminizes X. laevis calls into question the herbicide's role in that decline.' A report written in Environmental Science and Technology (May 15, 2008) cites the independent work of researchers in Japan, who were unable to replicate Hayes' work. 'The scientists found no hermaphrodite frogs; no increase in aromatase as measured by aromatase mRNA induction; and no increase in vitellogenin, another marker of feminization.' |
|---|
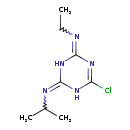
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-Chloro-4-(ethylamino)-6-(isopropylamino)-1,3,5-triazine | ChEBI | | 2-Chloro-4-ethylamino-6-isopropylamino-1,3,5-triazine | ChEBI | | 2-Chloro-4-ethylamino-6-isopropylamino-S-triazine | ChEBI | | 2-CHLORO-4-isopropylamino-6-ethylamino-1,3,5-triazine | ChEBI | | 2-Ethylamino-4-isopropylamino-6-chloro-S-triazine | ChEBI | | 6-Chloro-N-ethyl-n'-(1-methylethyl)-1,3,5-triazine-2,4-diamine | ChEBI | | 6-Chloro-N-ethyl-n'-isopropyl-1,3,5-triazine-2,4-diamine | ChEBI | | 1-Chloro-3-(ethylamino)-5-(isopropylamino)-2,4,6-triazine | HMDB | | 1-Chloro-3-(ethylamino)-5-(isopropylamino)-S-triazine | HMDB | | 2-Aethylamino-4-chlor-6-isopropylamino-1,3,5-triazin | HMDB | | 2-Chloro-4-(2-propylamino)-6-(ethylamino)-S-triazine | HMDB | | 2-Chloro-4-(ethylamino)-6-(isopropylamino)-S-triazine | HMDB | | 2-Chloro-4-ethylamineisopropylamine-S-triazine | HMDB | | 2-Chloro-4-isopropylamino-6-ethylamino -1,3,5-triazine | HMDB | | 6-Chloro-4-(ethylamino)-2-(isopropylamino)-S-triazine | HMDB | | 6-Chloro-N-ethyl-n'-(propan-2-yl)-1,3,5-triazine-2,4-diamine | HMDB | | Aatram | HMDB | | Aatrex | HMDB | | Aatrex 4l | HMDB | | Aatrex 4LC | HMDB | | Aatrex 80W | HMDB | | Aatrex nine-O | HMDB | | Actinite PK | HMDB | | Akticon | HMDB | | Aktikon | HMDB | | Aktikon PK | HMDB | | Aktinit a | HMDB | | Aktinit PK | HMDB | | Argezin | HMDB | | Atazinax | HMDB | | Atraflow | HMDB | | Atraflow plus | HMDB | | Atranex | HMDB | | Atrasine | HMDB | | Atrataf | HMDB | | Atratol | HMDB | | Atratol a | HMDB | | Atrazin | HMDB | | Atrazine 4l | HMDB | | Atrazine 80W | HMDB | | Atred | HMDB | | Atrex | HMDB | | Attrex | HMDB | | ATZ | HMDB | | Azinotox 500 | HMDB | | Candex | HMDB | | Cekuzina-T | HMDB | | Chromozin | HMDB | | Crisamina | HMDB | | Crisatrina | HMDB | | Crisazine | HMDB | | Cyazin | HMDB | | Farmco atrazine | HMDB | | Farmozine | HMDB | | Fenamin | HMDB | | Fenamine | HMDB | | Fenatrol | HMDB | | Fogard | HMDB | | Gesaprim | HMDB | | Gesaprim 50 | HMDB | | Gesoprim | HMDB | | Griffex | HMDB | | Griffex 4l | HMDB | | Herbatoxol | HMDB | | Hungazin | HMDB | | Hungazin PK | HMDB | | Inakor | HMDB | | Laddock | HMDB | | Maizina | HMDB | | Mebazine | HMDB | | Oleogesaprim | HMDB | | Pitezin | HMDB | | Primatol | HMDB | | Primatol a | HMDB | | Primaze | HMDB | | Primoleo | HMDB | | Radazin | HMDB | | Radazin T | HMDB | | Radizin | HMDB | | Radizine | HMDB | | Strazine | HMDB | | Triazine a 1294 | HMDB | | Vectal | HMDB | | Vectal SC | HMDB | | Weedex a | HMDB | | Wonuk | HMDB | | Zeaphos | HMDB | | Zeapos | HMDB | | Zeazin | HMDB | | Zeazine | HMDB | | Zeopos | HMDB | | Gesamprim | HMDB |
|
|---|
| Chemical Formula | C8H14ClN5 |
|---|
| Average Molecular Weight | 215.683 |
|---|
| Monoisotopic Molecular Weight | 215.09377318 |
|---|
| IUPAC Name | 6-chloro-N2-ethyl-N4-(propan-2-yl)-1,3,5-triazine-2,4-diamine |
|---|
| Traditional Name | atrazine |
|---|
| CAS Registry Number | 1912-24-9 |
|---|
| SMILES | CCN=C1NC(NC(Cl)=N1)=NC(C)C |
|---|
| InChI Identifier | InChI=1S/C8H14ClN5/c1-4-10-7-12-6(9)13-8(14-7)11-5(2)3/h5H,4H2,1-3H3,(H2,10,11,12,13,14) |
|---|
| InChI Key | MXWJVTOOROXGIU-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as chloro-s-triazines. These are aromatic compounds containing a 1,3,5-triazine ring that is substituted at the 2-position with a chlorine atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Triazines |
|---|
| Sub Class | 1,3,5-triazines |
|---|
| Direct Parent | Chloro-s-triazines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Chloro-s-triazine
- Aryl halide
- Aryl chloride
- Heteroaromatic compound
- Azacycle
- Organic nitrogen compound
- Organopnictogen compound
- Hydrocarbon derivative
- Organonitrogen compound
- Organochloride
- Organohalogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected and Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0uxr-6690000000-583f3e9640ef44c789ce | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-Q (Non-derivatized) | splash10-0uxu-9650000000-8ed90c24a7dac8c0763f | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - EI-B (Non-derivatized) | splash10-0uxr-6690000000-583f3e9640ef44c789ce | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-Q (Non-derivatized) | splash10-0uxu-9650000000-8ed90c24a7dac8c0763f | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0udi-4930000000-a800c662430d89a999bf | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-014i-0090000000-1f8dc1708e21447c72a5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-00xr-0950000000-698ba6253f9249ac28da | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-00di-0900000000-f20aaabd2097e4fd3d64 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-007k-0900000000-5b2179fcabb41a9319e0 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-000t-0900000000-bb61e6f57dde5f72c357 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-00di-0900000000-b333a1dd57dddfe03e6f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-014i-0090000000-066208bd5cb9497fe53e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-014i-0190000000-dadd355493b3a581e39c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-00xr-0960000000-1af13a0d4df29165659f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-00di-2900000000-37f6b1aa187e630690db | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-0g4j-6900000000-b43d1386f51d229ee0f4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-0uxs-9700000000-2523140d0327bff497ff | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-014i-0090000000-066208bd5cb9497fe53e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-014i-0190000000-ad3a817f71f2c0654dd8 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-00xr-1960000000-6cd3e48811e8b25a7c65 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-00di-2900000000-99e6afb84401fc17d138 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-0g4j-5900000000-d4992767b3b313839a0d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-0uxs-9800000000-eed7a36ca3c28d5cead3 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-00di-0900000000-e72e1e890a493331145d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0690000000-729ca7a9ad2de01cfe64 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00di-1910000000-0146c181e5d03eb2b37e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0h37-9600000000-7de90ffbda7ec9663fbc | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-3910000000-e195d0c31e12e3e7a831 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-06rj-7920000000-d4fc9069fd78b8033e88 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-7900000000-000a24131372a074e030 | View in MoNA |
|---|
| MS | Mass Spectrum (Electron Ionization) | splash10-0uxr-8890000000-b28a10166e9ef8427050 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, CDCl3, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100.40 MHz, CDCl3, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|