| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2018-07-17 17:21:02 UTC |
|---|
| Update Date | 2020-04-22 15:51:40 UTC |
|---|
| BMDB ID | BMDB0062229 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Cysteinyl-Asparagine |
|---|
| Description | Cysteinyl-asparagine, also known as C-N dipeptide or cys-asn, belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. Cysteinyl-asparagine is possibly soluble (in water) and a very strong basic compound (based on its pKa). |
|---|
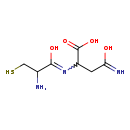
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| C-N Dipeptide | HMDB | | CN Dipeptide | HMDB | | Cys-asn | HMDB | | Cysteine asparagine dipeptide | HMDB | | Cysteine-asparagine dipeptide | HMDB | | Cysteinylasparagine | HMDB | | L-Cysteinyl-L-asparagine | HMDB | | 2-[(2-Amino-1-hydroxy-3-sulfanylpropylidene)amino]-3-(C-hydroxycarbonimidoyl)propanoate | HMDB | | 2-[(2-Amino-1-hydroxy-3-sulphanylpropylidene)amino]-3-(C-hydroxycarbonimidoyl)propanoate | HMDB | | 2-[(2-Amino-1-hydroxy-3-sulphanylpropylidene)amino]-3-(C-hydroxycarbonimidoyl)propanoic acid | HMDB |
|
|---|
| Chemical Formula | C7H13N3O4S |
|---|
| Average Molecular Weight | 235.261 |
|---|
| Monoisotopic Molecular Weight | 235.062676609 |
|---|
| IUPAC Name | 2-[(2-amino-1-hydroxy-3-sulfanylpropylidene)amino]-3-(C-hydroxycarbonimidoyl)propanoic acid |
|---|
| Traditional Name | 2-[(2-amino-1-hydroxy-3-sulfanylpropylidene)amino]-3-(C-hydroxycarbonimidoyl)propanoic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | NC(CS)C(O)=NC(CC(O)=N)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C7H13N3O4S/c8-3(2-15)6(12)10-4(7(13)14)1-5(9)11/h3-4,15H,1-2,8H2,(H2,9,11)(H,10,12)(H,13,14) |
|---|
| InChI Key | AYKQJQVWUYEZNU-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Dipeptides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-dipeptide
- Asparagine or derivatives
- N-acyl-alpha amino acid or derivatives
- N-acyl-alpha-amino acid
- Alpha-amino acid amide
- Cysteine or derivatives
- Alpha-amino acid or derivatives
- Fatty amide
- Fatty acid
- Fatty acyl
- Amino acid or derivatives
- Amino acid
- Primary carboxylic acid amide
- Carboxamide group
- Secondary carboxylic acid amide
- Monocarboxylic acid or derivatives
- Alkylthiol
- Carboxylic acid
- Organic nitrogen compound
- Organonitrogen compound
- Organooxygen compound
- Primary aliphatic amine
- Organosulfur compound
- Primary amine
- Hydrocarbon derivative
- Carbonyl group
- Organic oxide
- Amine
- Organopnictogen compound
- Organic oxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-002f-9500000000-db4f2dc1b9e86e0aa7ec | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0006-7940000000-4dc9c4ceb221166af6dc | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0170-3490000000-17bee52de00110ea2663 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004i-9310000000-dfc62ef252c063641b4b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a73-9100000000-b10645c1909673c5da82 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-1390000000-e336f74402a88c6c78ff | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000x-8940000000-e1f41694fb0b909ae826 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9100000000-e3c12eab9e76d8eaa663 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0190000000-1dd7b9e6c34a26e2704e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-004r-8950000000-12e8492ef8d11b008bcf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00bi-9100000000-415626ec168e41d32c14 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03gi-0910000000-4b2a44c3d9948b897fa5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03fr-2900000000-965b191618ad7c2a021e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9100000000-c846bb53d3d86322a329 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| General References | - Pan L, Yu J, Mi Z, Mo L, Jin H, Yao C, Ren D, Menghe B: A Metabolomics Approach Uncovers Differences between Traditional and Commercial Dairy Products in Buryatia (Russian Federation). Molecules. 2018 Mar 22;23(4). pii: molecules23040735. doi: 10.3390/molecules23040735. [PubMed:29565828 ]
|
|---|