| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-10-03 18:33:05 UTC |
|---|
| Update Date | 2020-04-22 15:48:42 UTC |
|---|
| BMDB ID | BMDB0012144 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
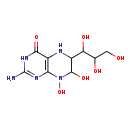
| Common Name | 2-Amino-4-oxo-6-(1',2',3'-trihydroxypropyl)-diquinoid-7,8-dihydroxypterin |
|---|
| Description | 2-Amino-4-oxo-6-(1',2',3'-trihydroxypropyl)-diquinoid-7,8-dihydroxypterin belongs to the class of organic compounds known as biopterins and derivatives. These are coenzymes containing a 2-amino-pteridine-4-one derivative. They are mainly synthesized in several parts of the body, including the pineal gland. Based on a literature review very few articles have been published on 2-Amino-4-oxo-6-(1',2',3'-trihydroxypropyl)-diquinoid-7,8-dihydroxypterin. |
|---|
| Structure | |
|---|
| Synonyms | Not Available |
|---|
| Chemical Formula | C9H15N5O6 |
|---|
| Average Molecular Weight | 289.2453 |
|---|
| Monoisotopic Molecular Weight | 289.102233237 |
|---|
| IUPAC Name | 2-amino-7,8-dihydroxy-6-(1,2,3-trihydroxypropyl)-3,4,5,6,7,8-hexahydropteridin-4-one |
|---|
| Traditional Name | 2-amino-7,8-dihydroxy-6-(1,2,3-trihydroxypropyl)-3,5,6,7-tetrahydropteridin-4-one |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | NC1=NC2=C(NC(C(O)C(O)CO)C(O)N2O)C(=O)N1 |
|---|
| InChI Identifier | InChI=1S/C9H15N5O6/c10-9-12-6-4(7(18)13-9)11-3(8(19)14(6)20)5(17)2(16)1-15/h2-3,5,8,11,15-17,19-20H,1H2,(H3,10,12,13,18) |
|---|
| InChI Key | KQZRTOGFDRQBSR-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as biopterins and derivatives. These are coenzymes containing a 2-amino-pteridine-4-one derivative. They are mainly synthesized in several parts of the body, including the pineal gland. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pteridines and derivatives |
|---|
| Sub Class | Pterins and derivatives |
|---|
| Direct Parent | Biopterins and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Biopterin
- Aminopyrimidine
- Pyrimidone
- Secondary aliphatic/aromatic amine
- Pyrimidine
- 1,3-aminoalcohol
- Heteroaromatic compound
- Vinylogous amide
- 1,2-aminoalcohol
- Secondary alcohol
- Alkanolamine
- Polyol
- Azacycle
- N-organohydroxylamine
- Secondary amine
- Primary alcohol
- Organic oxygen compound
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Organopnictogen compound
- Primary amine
- Hydrocarbon derivative
- Alcohol
- Organic nitrogen compound
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03k9-9250000000-ed65969b8a92432d02c1 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (4 TMS) - 70eV, Positive | splash10-0c00-3372190000-d873ed0eae62b6c36f12 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0090000000-8eb9a62dd0045b510707 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0fka-1690000000-9b5abf1f429a5406d1fa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-9330000000-d14d64770c6ed518d557 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-1090000000-62337634a8b5f1c81b1f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03ej-6950000000-0a26b118cfbd1118fc0e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-06tf-9310000000-43c935d8405e4bb829cf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0090000000-ed2094a9a02baf40342e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-06wd-1890000000-7d6a4aab5859324adf11 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-006x-7900000000-02a2fbdf086e417d02b6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00dl-0090000000-7d03a9ec97c10bef10d9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-0090000000-24421112d2d39eb45a56 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-003r-0910000000-0ecefc80926b16afa50e | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|