| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-10-03 18:22:14 UTC |
|---|
| Update Date | 2020-04-22 15:45:45 UTC |
|---|
| BMDB ID | BMDB0011648 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
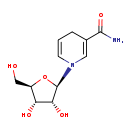
| Common Name | 1-(beta-D-Ribofuranosyl)-1,4-dihydronicotinamide |
|---|
| Description | 1-(beta-D-Ribofuranosyl)-1,4-dihydronicotinamide, also known as reduced nicotinamide riboside, belongs to the class of organic compounds known as glycosylamines. Glycosylamines are compounds consisting of an amine with a beta-N-glycosidic bond to a carbohydrate, thus forming a cyclic hemiaminal ether bond (alpha-amino ether). 1-(beta-D-Ribofuranosyl)-1,4-dihydronicotinamide is a strong basic compound (based on its pKa). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Reduced nicotinamide riboside | ChEBI | | 1-(b-D-Ribofuranosyl)-1,4-dihydronicotinamide | Generator | | 1-(Β-D-ribofuranosyl)-1,4-dihydronicotinamide | Generator |
|
|---|
| Chemical Formula | C11H16N2O5 |
|---|
| Average Molecular Weight | 256.2551 |
|---|
| Monoisotopic Molecular Weight | 256.105921632 |
|---|
| IUPAC Name | 1-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-1,4-dihydropyridine-3-carboxamide |
|---|
| Traditional Name | reduced nicotinamide riboside |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | NC(=O)C1=CN(C=CC1)[C@@H]1O[C@H](CO)[C@@H](O)[C@H]1O |
|---|
| InChI Identifier | InChI=1S/C11H16N2O5/c12-10(17)6-2-1-3-13(4-6)11-9(16)8(15)7(5-14)18-11/h1,3-4,7-9,11,14-16H,2,5H2,(H2,12,17)/t7-,8-,9-,11-/m1/s1 |
|---|
| InChI Key | MAKBMGXNXXXBFE-TURQNECASA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glycosylamines. Glycosylamines are compounds consisting of an amine with a beta-N-glycosidic bond to a carbohydrate, thus forming a cyclic hemiaminal ether bond (alpha-amino ether). |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Glycosylamines |
|---|
| Alternative Parents | |
|---|
| Substituents | - N-glycosyl compound
- N-substituted nicotinamide
- Pentose monosaccharide
- Dihydropyridine
- Hydropyridine
- Monosaccharide
- Vinylogous amide
- Tetrahydrofuran
- Carboxamide group
- Secondary alcohol
- Primary carboxylic acid amide
- Carboxylic acid derivative
- Enamine
- Oxacycle
- Azacycle
- Organoheterocyclic compound
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Carbonyl group
- Organic nitrogen compound
- Organonitrogen compound
- Alcohol
- Primary alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0a4r-9530000000-4d6ac885182221a2aa89 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-0a4i-6859800000-1a270761d5d36bc8a099 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4l-0190000000-bdc634b8a50f94b7870a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-6890000000-ab6e26f5e654c6811266 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-056r-6900000000-5629c3568948565f5b4e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-1290000000-49126d2627802aa8069d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00dr-2930000000-2f9086897c3a523ff340 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9200000000-d2c4dffcef36a3083d75 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0090000000-51f92fb4ea398fb5eb47 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0ac0-7910000000-75b65c4ba937a2b996e0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9400000000-f87d7c7a1d4c264a9b1f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-057i-1910000000-be783114ea0197565e8a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a7i-5970000000-d3c1ca153f39f41e5d41 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a59-9500000000-5a95eff6714627a2985b | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|