Showing metabocard for MG(0:0/14:1(9Z)/0:0) (BMDB0011531)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2016-10-03 18:20:11 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2020-05-11 19:18:12 UTC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BMDB ID | BMDB0011531 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
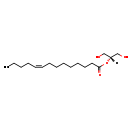
| Common Name | MG(0:0/14:1(9Z)/0:0) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | MG(0:0/14:1(9Z)/0:0) belongs to the family of monoradyglycerols, which are glycerolipids lipids containing a common glycerol backbone to which at one fatty acyl group is attached. Their general formula is [R1]OCC(CO[R2])O[R3]. MG(0:0/14:1(9Z)/0:0) is made up of one 9Z-tetradecenoyl(R2). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C17H32O4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight | 300.4336 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight | 300.230059512 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | 1,3-dihydroxypropan-2-yl (9Z)-tetradec-9-enoate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | 1,3-dihydroxypropan-2-yl (9Z)-tetradec-9-enoate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | [H]C(CO)(CO)OC(=O)CCCCCCC\C=C/CCCC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C17H32O4/c1-2-3-4-5-6-7-8-9-10-11-12-13-17(20)21-16(14-18)15-19/h5-6,16,18-19H,2-4,7-15H2,1H3/b6-5- | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | UETKPVCPWANBPE-WAYWQWQTSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as 2-monoacylglycerols. These are monoacylglycerols containing a glycerol acylated at the 2-position. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Lipids and lipid-like molecules | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Glycerolipids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Monoradylglycerols | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | 2-monoacylglycerols | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Ontology | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Expected but not Quantified | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofunction | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Application | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biospecimen Locations |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB0011531 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phenol Explorer Compound ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| FooDB ID | FDB028247 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KNApSAcK ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemspider ID | 24765768 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Compound ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BiGG ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| METLIN ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound | 53480959 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||