| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-10-03 18:14:01 UTC |
|---|
| Update Date | 2020-05-05 18:38:27 UTC |
|---|
| BMDB ID | BMDB0011170 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
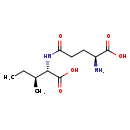
| Common Name | L-gamma-glutamyl-L-isoleucine |
|---|
| Description | gamma-Glutamylisoleucine, also known as γ-L-glutamyl-L-isoleucine or L-gamma-glu-L-ile, belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. Based on a literature review a significant number of articles have been published on gamma-Glutamylisoleucine. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| L-gamma-Glu-L-ile | ChEBI | | L-g-Glu-L-ile | Generator | | L-Γ-glu-L-ile | Generator | | g-Glutamylisoleucine | Generator | | Γ-glutamylisoleucine | Generator | | Γ-glu-ile | HMDB | | Γ-L-glu-L-ile | HMDB | | Γ-L-glutamyl-L-isoleucine | HMDB | | L-Γ-glutamyl-L-isoleucine | HMDB | | N-Γ-glutamylisoleucine | HMDB | | N-L-Γ-glutamylisoleucine | HMDB | | N-L-Γ-glutamyl-L-isoleucine | HMDB | | gamma-Glu-ile | HMDB | | gamma-L-Glu-L-ile | HMDB | | gamma-L-Glutamyl-L-isoleucine | HMDB | | L-gamma-Glutamyl-L-isoleucine | HMDB | | N-gamma-Glutamylisoleucine | HMDB | | N-L-gamma-Glutamylisoleucine | HMDB | | N-L-gamma-Glutamyl-L-isoleucine | HMDB | | g-Glu-ile | HMDB | | N-Γ-L-glutamyl-L-isoleucine | HMDB | | N-gamma-L-Glutamyl-L-isoleucine | HMDB | | gamma-Glutamylisoleucine | HMDB |
|
|---|
| Chemical Formula | C11H20N2O5 |
|---|
| Average Molecular Weight | 260.29 |
|---|
| Monoisotopic Molecular Weight | 260.137221752 |
|---|
| IUPAC Name | (2S,3S)-2-[(4S)-4-amino-4-carboxybutanamido]-3-methylpentanoic acid |
|---|
| Traditional Name | (2S,3S)-2-[(4S)-4-amino-4-carboxybutanamido]-3-methylpentanoic acid |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | CC[C@H](C)[C@H](NC(=O)CC[C@H](N)C(O)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C11H20N2O5/c1-3-6(2)9(11(17)18)13-8(14)5-4-7(12)10(15)16/h6-7,9H,3-5,12H2,1-2H3,(H,13,14)(H,15,16)(H,17,18)/t6-,7-,9-/m0/s1 |
|---|
| InChI Key | SNCKGJWJABDZHI-ZKWXMUAHSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as dipeptides. These are organic compounds containing a sequence of exactly two alpha-amino acids joined by a peptide bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Dipeptides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alpha-dipeptide
- Gamma-glutamyl alpha-amino acid
- Glutamine or derivatives
- Isoleucine or derivatives
- N-acyl-alpha-amino acid
- N-acyl-alpha amino acid or derivatives
- N-acyl-l-alpha-amino acid
- Alpha-amino acid
- Alpha-amino acid or derivatives
- L-alpha-amino acid
- Methyl-branched fatty acid
- Branched fatty acid
- N-acyl-amine
- Fatty acyl
- Fatty acid
- Dicarboxylic acid or derivatives
- Fatty amide
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Carboxamide group
- Amino acid
- Carboxylic acid
- Organic nitrogen compound
- Primary aliphatic amine
- Organonitrogen compound
- Organooxygen compound
- Primary amine
- Hydrocarbon derivative
- Carbonyl group
- Organic oxide
- Organopnictogen compound
- Amine
- Organic oxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Detected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014l-0490000000-9a8493d1209721c96c45 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0159-4940000000-c8d2557d4ece2b3e1f48 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a59-9400000000-c4806ce4eb33b070ec04 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0aor-0190000000-fba2af6da925fa69e695 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-05ng-2980000000-c4854314104d3f1dc06f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01x0-9500000000-3ebdef8b8d4284510598 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0290000000-37e1c45547c2f6b7c83f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-4930000000-656c949a22ffdbe66bb2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9100000000-91a811eea14294cf8bbf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0390000000-c75ac26b7fd31c08f923 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-1900000000-04a27ca349255e4db60b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9300000000-ef578d2b25c43747f2b8 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|