| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:24:20 UTC |
|---|
| Update Date | 2020-04-22 15:18:08 UTC |
|---|
| BMDB ID | BMDB0006620 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
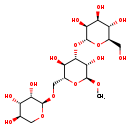
| Common Name | Fucosyllactose |
|---|
| Description | Fucosyllactose belongs to the class of organic compounds known as oligosaccharides. These are carbohydrates made up of 3 to 10 monosaccharide units linked to each other through glycosidic bonds. Based on a literature review very few articles have been published on Fucosyllactose. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| mono(6-Deoxy-L-galactopyranoside 4-O-beta-D-galactopyranosyl-D-glucose | HMDB | | mono(6-Deoxy-L-galactopyranoside 4-O-beta-delta-galactopyranosyl-delta-glucose | HMDB |
|
|---|
| Chemical Formula | C18H32O15 |
|---|
| Average Molecular Weight | 488.4377 |
|---|
| Monoisotopic Molecular Weight | 488.174120354 |
|---|
| IUPAC Name | (2R,3S,4S,5S,6R)-2-{[(2S,3S,4S,5R,6R)-3,5-dihydroxy-2-methoxy-6-({[(2R,3S,4S,5R)-3,4,5-trihydroxyoxan-2-yl]oxy}methyl)oxan-4-yl]oxy}-6-(hydroxymethyl)oxane-3,4,5-triol |
|---|
| Traditional Name | fucosyllactose |
|---|
| CAS Registry Number | 108795-32-0 |
|---|
| SMILES | CO[C@H]1O[C@H](CO[C@H]2OC[C@@H](O)[C@H](O)[C@@H]2O)[C@@H](O)[C@H](O[C@H]2O[C@H](CO)[C@@H](O)[C@H](O)[C@@H]2O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C18H32O15/c1-28-16-14(27)15(33-18-13(26)11(24)9(22)6(2-19)31-18)10(23)7(32-16)4-30-17-12(25)8(21)5(20)3-29-17/h5-27H,2-4H2,1H3/t5-,6-,7-,8+,9-,10-,11+,12+,13+,14+,15+,16+,17-,18-/m1/s1 |
|---|
| InChI Key | RTVRUWIBAVHRQX-PMEZUWKYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as oligosaccharides. These are carbohydrates made up of 3 to 10 monosaccharide units linked to each other through glycosidic bonds. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Oligosaccharides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Oligosaccharide
- O-glycosyl compound
- Glycosyl compound
- Oxane
- Secondary alcohol
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Acetal
- Hydrocarbon derivative
- Primary alcohol
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00di-5524900000-a34f1e16493a78c7e433 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-03yi-3701219000-29ca3f06f6936c51d11f | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_3_29) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS ("Fucosyllactose,3TBDMS,#29" TMS) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0adr-0209700000-21c1a7e0e925ddaeb112 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a6u-0809000000-2a50f0a23e176a847de2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-056u-1914000000-8e09b33d9db2d27db6f7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002r-2716900000-ac5fc4ebf2fc09562fea | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00pj-2924200000-e7c15c37365682db02ad | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-002f-7941000000-3bbe969391c324f9a1e4 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0002900000-0bebd00ce2822ebbcdb7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0btc-6207900000-7ccfab956dce719d6734 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9411000000-8b51e6e270c89dcc4582 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0205900000-1956792dd589e71853d8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-054n-0925200000-8ceb756d5dbadd648af5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-07vi-4930000000-a933094b772ab1a06564 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|