| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:20:07 UTC |
|---|
| Update Date | 2020-04-22 15:16:50 UTC |
|---|
| BMDB ID | BMDB0006083 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Troxerutin |
|---|
| Description | Troxerutin, also known as venoruton or veniten retard, belongs to the class of organic compounds known as flavonoid-3-o-glycosides. These are phenolic compounds containing a flavonoid moiety which is O-glycosidically linked to carbohydrate moiety at the C3-position. Based on a literature review a significant number of articles have been published on Troxerutin. |
|---|
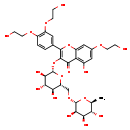
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Trioxyethylrutin | HMDB | | Vitamin P4 | HMDB | | Flavonoid | HMDB | | 3',4',7-Tri-O-(b-hydroxyethyl)rutoside | HMDB | | 3',4',7-Tris(hydroxyethyl)rutin | HMDB | | 3,5-Dihydroxy-3',4',7-tris(2-hydroxyethoxy)flavone 3-rutinoside | HMDB | | 3-[6-O-(6-Deoxy-a-L-mannopyranosyl)-b-D-glucopyranoside]3,5-dihydroxy-3',4',7-tris(2-hydroxyethoxy)-flavone | HMDB | | 3-[6-O-(6-Deoxy-alpha-L-mannopyranosyl)-beta-delta-glucopyranoside]3,5-dihydroxy-3',4',7-tris(2-hydroxyethoxy)-flavone | HMDB | | Factor p-zyma | HMDB | | Posorutin | HMDB | | Rufen-P4 | HMDB | | Ruven | HMDB | | THR | HMDB | | TriHER | HMDB | | Tris(hydroxyethyl)rutin | HMDB | | Tris(hydroxyethyl)rutoside | HMDB | | Tris-O-(2-hydroxyethyl)rutin | HMDB | | Tris-O-(b-hydroxyethyl)rutoside | HMDB | | Troxerutine | HMDB | | Vastribil | HMDB | | Veinamitol | HMDB | | Veniten | HMDB | | Venoruton P4 | HMDB | | Z 6000 | HMDB | | Venorutin | HMDB | | Venotrulan trox | HMDB | | 3',4',7-Tris(O-(2- hydroxyethyl))rutin | HMDB | | Troxerutin-ratiopharm | HMDB | | Oxerutin | HMDB | | Veno SL | HMDB | | Paroven | HMDB | | Relvene | HMDB | | Rhéoflux | HMDB | | 3',4',7-Trihydroxyethylrutin | HMDB | | Teboven | HMDB | | Troxérutine mazal | HMDB | | Veniten retard | HMDB | | Venoruton | HMDB | | Troxeven | HMDB |
|
|---|
| Chemical Formula | C33H42O19 |
|---|
| Average Molecular Weight | 742.6752 |
|---|
| Monoisotopic Molecular Weight | 742.232029162 |
|---|
| IUPAC Name | 2-[3,4-bis(2-hydroxyethoxy)phenyl]-5-hydroxy-7-(2-hydroxyethoxy)-3-{[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-({[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxy}methyl)oxan-2-yl]oxy}-4H-chromen-4-one |
|---|
| Traditional Name | ruven |
|---|
| CAS Registry Number | 7085-55-4 |
|---|
| SMILES | C[C@@H]1O[C@@H](OC[C@H]2O[C@@H](OC3=C(OC4=CC(OCCO)=CC(O)=C4C3=O)C3=CC(OCCO)=C(OCCO)C=C3)[C@H](O)[C@@H](O)[C@@H]2O)[C@H](O)[C@H](O)[C@H]1O |
|---|
| InChI Identifier | InChI=1S/C33H42O19/c1-14-23(38)26(41)28(43)32(49-14)48-13-21-24(39)27(42)29(44)33(51-21)52-31-25(40)22-17(37)11-16(45-7-4-34)12-20(22)50-30(31)15-2-3-18(46-8-5-35)19(10-15)47-9-6-36/h2-3,10-12,14,21,23-24,26-29,32-39,41-44H,4-9,13H2,1H3/t14-,21+,23-,24+,26+,27-,28+,29+,32+,33-/m0/s1 |
|---|
| InChI Key | IYVFNTXFRYQLRP-VVSTWUKXSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as flavonoid-3-o-glycosides. These are phenolic compounds containing a flavonoid moiety which is O-glycosidically linked to carbohydrate moiety at the C3-position. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Phenylpropanoids and polyketides |
|---|
| Class | Flavonoids |
|---|
| Sub Class | Flavonoid glycosides |
|---|
| Direct Parent | Flavonoid-3-O-glycosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - Flavonoid-3-o-glycoside
- 5-hydroxyflavonoid
- Hydroxyflavonoid
- Flavone
- O-glycosyl compound
- Glycosyl compound
- Chromone
- Disaccharide
- Benzopyran
- 1-benzopyran
- Phenoxy compound
- Phenol ether
- 1-hydroxy-4-unsubstituted benzenoid
- 1-hydroxy-2-unsubstituted benzenoid
- Alkyl aryl ether
- Pyranone
- Oxane
- Pyran
- Monocyclic benzene moiety
- Benzenoid
- Heteroaromatic compound
- Vinylogous acid
- Secondary alcohol
- Acetal
- Ether
- Organoheterocyclic compound
- Oxacycle
- Polyol
- Primary alcohol
- Organic oxygen compound
- Hydrocarbon derivative
- Organic oxide
- Alcohol
- Organooxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 181 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_7) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_8) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_9) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_10) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_11) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_12) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_13) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_14) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_15) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-002r-1103914700-9118812e6a139677e7ec | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00kr-1204920000-e1a6fe258249bdf38cca | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0309413000-18cde334d33eb7397b73 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-009w-3311738900-bc8203f2a87b41de15c6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01yk-2407319300-d09ced6a80acfeabd4f6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0019-1309403000-dcb1e6055caa3e58fcf0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0000900200-67645d791629add48e0c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000o-0000900900-a2300754ed17d9d7b991 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-0000900000-8d96ed536fe3359a7ffa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0000000900-acfa986c3ddec08dd01a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000x-0000500900-7ece5d3b366c14d1584c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-0010900100-c1e52788f916928bee37 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|