| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 23:04:46 UTC |
|---|
| Update Date | 2020-04-22 15:12:20 UTC |
|---|
| BMDB ID | BMDB0003454 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 4-Pyridoxolactone |
|---|
| Description | 4-Pyridoxolactone belongs to the class of organic compounds known as pyridinecarboxylic acids. Pyridinecarboxylic acids are compounds containing a pyridine ring bearing a carboxylic acid group. 4-Pyridoxolactone exists in all eukaryotes, ranging from yeast to plants to humans. Based on a literature review a significant number of articles have been published on 4-Pyridoxolactone. |
|---|
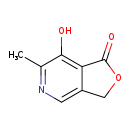
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 4-Pyridoxic acid lactone | ChEBI | | 4-Pyridoxate lactone | Generator | | beta-Pyracin | HMDB |
|
|---|
| Chemical Formula | C8H7NO3 |
|---|
| Average Molecular Weight | 165.1461 |
|---|
| Monoisotopic Molecular Weight | 165.042593095 |
|---|
| IUPAC Name | 7-hydroxy-6-methyl-1H,3H-furo[3,4-c]pyridin-1-one |
|---|
| Traditional Name | 4-pyridoxolactone |
|---|
| CAS Registry Number | 4753-19-9 |
|---|
| SMILES | CC1=NC=C2COC(=O)C2=C1O |
|---|
| InChI Identifier | InChI=1S/C8H7NO3/c1-4-7(10)6-5(2-9-4)3-12-8(6)11/h2,10H,3H2,1H3 |
|---|
| InChI Key | HHPDVQLBYQFYFA-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyridinecarboxylic acids. Pyridinecarboxylic acids are compounds containing a pyridine ring bearing a carboxylic acid group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pyridines and derivatives |
|---|
| Sub Class | Pyridinecarboxylic acids and derivatives |
|---|
| Direct Parent | Pyridinecarboxylic acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyridine carboxylic acid
- Methylpyridine
- Hydroxypyridine
- Vinylogous acid
- Heteroaromatic compound
- Carboxylic acid ester
- Lactone
- Carboxylic acid derivative
- Monocarboxylic acid or derivatives
- Azacycle
- Oxacycle
- Organopnictogen compound
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Organic nitrogen compound
- Organic oxygen compound
- Hydrocarbon derivative
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-00ri-2900000000-5f6a91d1eeb5eaa49adc | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-00dl-5950000000-11ff12c2dd36530c4236 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0900000000-aa1266c816944909b39a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-0900000000-d0258c632349f5e836e0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00dj-7900000000-30cd1dadf39f33995b09 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0900000000-3fd21d96beb6c70780d6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-0900000000-2b177a3586d7e6d68836 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00e9-9800000000-3abeb7516d954dda95b1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-0900000000-65490e650f013cfe115e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03k9-0900000000-7ece33897451c1af0c35 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4v-7900000000-3ee7b8f8ce87fd24b311 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014i-0900000000-52879b1c24e4e827f43a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-0900000000-1dc0c584a9bf4ad25064 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0072-5900000000-8631bb6ae260af7ebb9b | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|