| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:47:59 UTC |
|---|
| Update Date | 2020-04-22 15:08:48 UTC |
|---|
| BMDB ID | BMDB0001786 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Ethenodeoxyadenosine |
|---|
| Description | Ethenodeoxyadenosine belongs to the class of organic compounds known as purines and purine derivatives. These are aromatic heterocyclic compounds containing a purine moiety, which is formed a pyrimidine-ring ring fused to an imidazole ring. Based on a literature review a small amount of articles have been published on Ethenodeoxyadenosine. |
|---|
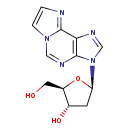
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,N(6)-Deoxyethenoadenosine | MeSH | | 1,N(6)-Ethenodeoxyadenosine | MeSH | | 1,N2-Etheno-2'-deoxyguanosine | MeSH | | 1,N6-Etheno-2'-deoxyadenosine | MeSH | | 2'-Deoxy-2-deuteroethenoadenosine | MeSH | | 1,N6-etheno-DA | HMDB | | 1,N6-Ethenodeoxyadenosine | HMDB | | N1,N6-etheno-2'-Deoxyadenosine | HMDB |
|
|---|
| Chemical Formula | C12H13N5O3 |
|---|
| Average Molecular Weight | 275.2633 |
|---|
| Monoisotopic Molecular Weight | 275.101839307 |
|---|
| IUPAC Name | (2R,3S,5R)-2-(hydroxymethyl)-5-{3H-imidazo[2,1-f]purin-3-yl}oxolan-3-ol |
|---|
| Traditional Name | ethenodeoxyadenosine |
|---|
| CAS Registry Number | 68498-25-9 |
|---|
| SMILES | OC[C@H]1O[C@H](C[C@@H]1O)N1C=NC2=C1N=CN1C=CN=C21 |
|---|
| InChI Identifier | InChI=1S/C12H13N5O3/c18-4-8-7(19)3-9(20-8)17-6-14-10-11-13-1-2-16(11)5-15-12(10)17/h1-2,5-9,18-19H,3-4H2/t7-,8+,9+/m0/s1 |
|---|
| InChI Key | XQQIMTUYVDUWKJ-DJLDLDEBSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as purines and purine derivatives. These are aromatic heterocyclic compounds containing a purine moiety, which is formed a pyrimidine-ring ring fused to an imidazole ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | Purines and purine derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Purine
- N-substituted imidazole
- Pyrimidine
- Heteroaromatic compound
- Tetrahydrofuran
- Imidazole
- Azole
- Secondary alcohol
- Oxacycle
- Azacycle
- Hydrocarbon derivative
- Organic nitrogen compound
- Primary alcohol
- Organooxygen compound
- Organonitrogen compound
- Alcohol
- Organopnictogen compound
- Organic oxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-054o-9170000000-59787ef95e13965a6f74 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-0v4i-9855100000-1aa19e03cd5abf43bec2 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-004i-0090000000-1cbcf897b8d539f74c9d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-03di-0900000000-7351b3ecaf69532e4ae9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-03di-1900000000-cea99433c51ea3b57b8c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0920000000-4723a3e7bccfd3fb82a3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0900000000-319e0450fd20aa8b295f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-0900000000-123c329a87834ac8c719 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0290000000-8d53e894625170dfa26f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0910000000-c8a7d5a2ae45462ace75 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-0900000000-944674f1809414d8aa98 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0900000000-491bffd5eb2009008d64 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-0900000000-491bffd5eb2009008d64 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-0900000000-d0145665222b050d7b7c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0190000000-bf4918a4b52f776c05be | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0900000000-9997b969fd66d57e3889 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-0900000000-614f89543dcac00da239 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, 5%_DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, 5%_DMSO, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|