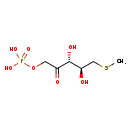| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:43:46 UTC |
|---|
| Update Date | 2020-04-22 15:07:31 UTC |
|---|
| BMDB ID | BMDB0001299 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 5-Methylthioribulose 1-phosphate |
|---|
| Description | 5-Methylthioribulose 1-phosphate, also known as 1PMT-ribulose or 1-phospho-5-S-methylthioribulose, belongs to the class of organic compounds known as glycerone phosphates. These are organic compounds containing a glycerone moiety that carries a phosphate group at the O-1 or O-2 position. 5-Methylthioribulose 1-phosphate is an extremely weak basic (essentially neutral) compound (based on its pKa). 5-Methylthioribulose 1-phosphate exists in all living species, ranging from bacteria to humans. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 5-S-Methyl-1-O-phosphono-5-thio-D-ribulose | ChEBI | | 5-(Methylsulfanyl)-D-ribulose 1-phosphate | Kegg | | 5-(Methylsulfanyl)-D-ribulose 1-phosphoric acid | Generator | | 5-(Methylsulphanyl)-D-ribulose 1-phosphate | Generator | | 5-(Methylsulphanyl)-D-ribulose 1-phosphoric acid | Generator | | 5-Methylthioribulose 1-phosphoric acid | Generator | | 1-Phospho-5-S-methylthioribulose | HMDB | | 1-Phosphomethylthioribulose | HMDB | | 1PMT-Ribulose | HMDB | | 5-Methylthio-2-ribulose-1-phosphate | HMDB | | 5-Methylthio-5-deoxy-D-ribulose 1-phosphate | HMDB | | 5-Methylthio-5-deoxy-D-ribulose-1-phosphate | HMDB | | 5-Methylthioribulose-1-phosphate | HMDB | | 5-S-Methyl-5-thio-D-erythro-pent-2-ulose 1-(dihydrogen phosphate) | HMDB | | 5-S-Methyl-5-thio-D-ribulose 1-(dihydrogen phosphate) | HMDB | | Methylthioribulose-1-phosphate | HMDB | | MTRu-1-p | HMDB | | S-Methyl-5-thio-D-ribulose 1-phosphate | HMDB | | 1-PMT-Ribulose | HMDB | | Methylthioribulose 1-phosphate | HMDB | | 5-(Methylthio)ribulose 1-phosphate | HMDB |
|
|---|
| Chemical Formula | C6H13O7PS |
|---|
| Average Molecular Weight | 260.202 |
|---|
| Monoisotopic Molecular Weight | 260.011959972 |
|---|
| IUPAC Name | {[(3R,4S)-3,4-dihydroxy-5-(methylsulfanyl)-2-oxopentyl]oxy}phosphonic acid |
|---|
| Traditional Name | methylthioribulose-1-phosphate |
|---|
| CAS Registry Number | 86316-83-8 |
|---|
| SMILES | CSC[C@@H](O)[C@@H](O)C(=O)COP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H13O7PS/c1-15-3-5(8)6(9)4(7)2-13-14(10,11)12/h5-6,8-9H,2-3H2,1H3,(H2,10,11,12)/t5-,6+/m1/s1 |
|---|
| InChI Key | CNSJRYUMVMWNMC-RITPCOANSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as glycerone phosphates. These are organic compounds containing a glycerone moiety that carries a phosphate group at the O-1 or O-2 position. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbonyl compounds |
|---|
| Direct Parent | Glycerone phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Glycerone phosphate
- Monoalkyl phosphate
- Acyloin
- Beta-hydroxy ketone
- Monosaccharide
- Organic phosphoric acid derivative
- Phosphoric acid ester
- Alkyl phosphate
- Alpha-hydroxy ketone
- Secondary alcohol
- 1,2-diol
- Thioether
- Sulfenyl compound
- Dialkylthioether
- Organosulfur compound
- Alcohol
- Organic oxide
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01w1-9710000000-da3fc8534da2cd8957f4 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-03di-7973000000-51639c96fd3e70d14bf8 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03dl-3590000000-48a371cabc865281cedb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01pc-3920000000-0f483ba04442297b4622 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000g-9300000000-6e02d97a8ab3c704f801 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-9310000000-4f7b1426b6d9f8a88ffa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004j-9100000000-1e987229b4853fb45072 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-fe6a484561738ead9cd1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-1590000000-1113571c4892aa6f836e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03ka-9400000000-da5dcbcf9b27c541867d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-9000000000-fec7f320592a6e9bc867 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4r-3490000000-1b9248527c15132ff9ee | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004j-9000000000-f420c16db721baadbf7c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-06c27ade5d32e8e9acda | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| Synthesis Reference | Imker, Heidi J.; Fedorov, Alexander A.; Fedorov, Elena V.; Almo, Steven C.; Gerlt, John A. Mechanistic Diversity in the RuBisCO Superfamily: The "Enolase" in the Methionine Salvage Pathway in Geobacillus kaustophilus. Biochemistry (2007), 46(13), 4077-4089. |
|---|