| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:42:10 UTC |
|---|
| Update Date | 2020-04-22 15:07:00 UTC |
|---|
| BMDB ID | BMDB0001182 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 6,8-Dihydroxypurine |
|---|
| Description | 6,8-Dihydroxypurine, also known as 6,8-purinediol or 8-oxohypoxanthine, belongs to the class of organic compounds known as hypoxanthines. Hypoxanthines are compounds containing the purine derivative 1H-purin-6(9H)-one. Purine is a bicyclic aromatic compound made up of a pyrimidine ring fused to an imidazole ring. Based on a literature review a significant number of articles have been published on 6,8-Dihydroxypurine. |
|---|
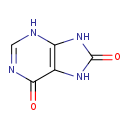
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 6,8-Purinediol | HMDB | | 7,9-Dihydro-1H-purine-6,8-dione | HMDB | | 7,9-Dihydro-8H-purin-8-one | HMDB | | 8-Oxohypoxanthine | HMDB | | Purine-6,8(1H,9H)-dione | HMDB | | Purine-6,8-diol | HMDB | | Purine-6,8-dione | HMDB |
|
|---|
| Chemical Formula | C5H4N4O2 |
|---|
| Average Molecular Weight | 152.1109 |
|---|
| Monoisotopic Molecular Weight | 152.033425392 |
|---|
| IUPAC Name | 6,7,8,9-tetrahydro-3H-purine-6,8-dione |
|---|
| Traditional Name | 7,9-dihydro-3H-purine-6,8-dione |
|---|
| CAS Registry Number | 13231-00-0 |
|---|
| SMILES | O=C1NC2=C(N1)C(=O)N=CN2 |
|---|
| InChI Identifier | InChI=1S/C5H4N4O2/c10-4-2-3(6-1-7-4)9-5(11)8-2/h1H,(H3,6,7,8,9,10,11) |
|---|
| InChI Key | BYUOBSUZYQAFJM-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as hypoxanthines. Hypoxanthines are compounds containing the purine derivative 1H-purin-6(9H)-one. Purine is a bicyclic aromatic compound made up of a pyrimidine ring fused to an imidazole ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | Hypoxanthines |
|---|
| Alternative Parents | |
|---|
| Substituents | - 6-oxopurine
- Hypoxanthine
- Pyrimidone
- Pyrimidine
- Azole
- Imidazole
- Heteroaromatic compound
- Vinylogous amide
- Urea
- Azacycle
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 0.167 mg/mL at 20 °C | Not Available | | LogP | 0.246 | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0zi1-6900000000-b597900216a7c18ff4ec | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900000000-73408e2a019cf6145389 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0900000000-4ec7e27bd75758d5243d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000x-9400000000-4b36792f2a9b31c0e39f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0900000000-0c74ea3af474d10b7ea8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-0900000000-30080f0e6c244018af86 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-053u-9600000000-23699c571e13d9a8c982 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900000000-ac450658003af6c0a109 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0900000000-00ec07b7bda30887c327 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-05ai-9200000000-f8c6511273a5bfd31f74 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0900000000-757517510c89a3191171 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0fk9-2900000000-b1f56627b7499b83ee76 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-5b696d1d5422470c88ea | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| Synthesis Reference | Ohtsuka, Yozo; Sugimoto, Kikuo. Oxazolopyrimidines. III. Formation of 6,8-dihydroxypurine by the oxidation of 7-aminooxazolo[5,4-d]pyrimidine with hydrogen peroxide-acetic acid. Bulletin of the Chemical Society of Japan (1970), 43(7), 2281. |
|---|