| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:38:59 UTC |
|---|
| Update Date | 2020-05-11 20:50:44 UTC |
|---|
| BMDB ID | BMDB0000946 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
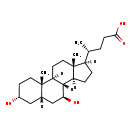
| Common Name | Ursodeoxycholic acid |
|---|
| Description | Ursodeoxycholic acid, also known as ursodeoxycholate or actigall, belongs to the class of organic compounds known as dihydroxy bile acids, alcohols and derivatives. Dihydroxy bile acids, alcohols and derivatives are compounds containing or derived from a bile acid or alcohol, and which bears exactly two carboxylic acid groups. Ursodeoxycholic acid is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (3alpha,5beta,7beta)-3,7-Dihydroxycholan-24-Oic acid | ChEBI | | 3alpha,7beta-Dihydroxy-5beta-cholan-24-Oic acid | ChEBI | | Actigall | ChEBI | | Ursodeoxycholate | ChEBI | | Ursodiol | ChEBI | | Urso | Kegg | | (3a,5b,7b)-3,7-Dihydroxycholan-24-Oate | Generator | | (3a,5b,7b)-3,7-Dihydroxycholan-24-Oic acid | Generator | | (3alpha,5beta,7beta)-3,7-Dihydroxycholan-24-Oate | Generator | | (3Α,5β,7β)-3,7-dihydroxycholan-24-Oate | Generator | | (3Α,5β,7β)-3,7-dihydroxycholan-24-Oic acid | Generator | | 3a,7b-Dihydroxy-5b-cholan-24-Oate | Generator | | 3a,7b-Dihydroxy-5b-cholan-24-Oic acid | Generator | | 3alpha,7beta-Dihydroxy-5beta-cholan-24-Oate | Generator | | 3Α,7β-dihydroxy-5β-cholan-24-Oate | Generator | | 3Α,7β-dihydroxy-5β-cholan-24-Oic acid | Generator | | 3 alpha,7 beta-Dihydroxy-5 beta-cholan-24-Oic acid | HMDB | | Acid, deoxyursocholic | HMDB | | Acid, ursacholic | HMDB | | Acid, ursodeoxycholic | HMDB | | Cholit-ursan | HMDB | | Cholofalk | HMDB | | Delursan | HMDB | | Deoxyursocholic acid | HMDB | | Ursochol | HMDB | | Ursogal | HMDB | | Ursolite | HMDB | | Ursolvan | HMDB | | Urso heumann | HMDB | | Ursobilane | HMDB | | Ursofalk | HMDB | | Destolit | HMDB | | Sodium ursodeoxycholate | HMDB | | Ursacholic acid | HMDB | | Ursodeoxycholate, sodium | HMDB | | 3 alpha,7 beta Dihydroxy 5 beta cholan 24 Oic acid | HMDB | | Urdox | HMDB | | (3a,5b,7b)-3,7-Dihydroxy-cholan-24-Oate | HMDB | | (3a,5b,7b)-3,7-Dihydroxy-cholan-24-Oic acid | HMDB | | 3,7-Dihydroxycholan-24-Oic acid | HMDB | | 3-alpha,7-beta-Dihydroxy-5-beta-cholanoic acid | HMDB | | 3-alpha,7-beta-Dihydroxycholanic acid | HMDB | | 3-alpha,7-beta-Dioxycholanic acid | HMDB | | Antigall | HMDB | | Urosdesoxycholate | HMDB | | Urosdesoxycholic acid | HMDB | | Ursodeoxycholicacid | HMDB | | Ursodexycholate | HMDB | | Ursodexycholic acid | HMDB | | Aventis brand OF ursodeoxycholic acid | HMDB | | Axcan brand OF ursodeoxycholic acid | HMDB | | Falk brand OF ursodeoxycholic acid | HMDB | | Farmasa brand OF ursodeoxycholic acid | HMDB | | Galen brand OF ursodeoxycholic acid | HMDB | | Orphan brand OF ursodeoxycholic acid | HMDB | | Antigen brand OF ursodeoxycholic acid | HMDB | | CP Brand OF ursodeoxycholic acid | HMDB | | Heumann brand OF ursodeoxycholic acid | HMDB | | Niddapharm brand OF ursodeoxycholic acid | HMDB | | Norgine brand OF ursodeoxycholic acid | HMDB | | Sanofi synthelabo brand OF ursodeoxycholic acid | HMDB | | Estedi brand OF ursodeoxycholic acid | HMDB | | Provalis brand OF ursodeoxycholic acid | HMDB | | Tramedico brand OF ursodeoxycholic acid | HMDB | | Vita brand OF ursodeoxycholic acid | HMDB | | Zambon brand OF ursodeoxycholic acid | HMDB |
|
|---|
| Chemical Formula | C24H40O4 |
|---|
| Average Molecular Weight | 392.572 |
|---|
| Monoisotopic Molecular Weight | 392.292659768 |
|---|
| IUPAC Name | (4R)-4-[(1S,2S,5R,7S,9S,10R,11S,14R,15R)-5,9-dihydroxy-2,15-dimethyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadecan-14-yl]pentanoic acid |
|---|
| Traditional Name | (4R)-4-[(1S,2S,5R,7S,9S,10R,11S,14R,15R)-5,9-dihydroxy-2,15-dimethyltetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadecan-14-yl]pentanoic acid |
|---|
| CAS Registry Number | 128-13-2 |
|---|
| SMILES | [H][C@@]1(CC[C@@]2([H])[C@]3([H])[C@@H](O)C[C@]4([H])C[C@H](O)CC[C@]4(C)[C@@]3([H])CC[C@]12C)[C@H](C)CCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C24H40O4/c1-14(4-7-21(27)28)17-5-6-18-22-19(9-11-24(17,18)3)23(2)10-8-16(25)12-15(23)13-20(22)26/h14-20,22,25-26H,4-13H2,1-3H3,(H,27,28)/t14-,15+,16-,17-,18+,19+,20+,22+,23+,24-/m1/s1 |
|---|
| InChI Key | RUDATBOHQWOJDD-UZVSRGJWSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as dihydroxy bile acids, alcohols and derivatives. Dihydroxy bile acids, alcohols and derivatives are compounds containing or derived from a bile acid or alcohol, and which bears exactly two carboxylic acid groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Bile acids, alcohols and derivatives |
|---|
| Direct Parent | Dihydroxy bile acids, alcohols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Dihydroxy bile acid, alcohol, or derivatives
- 3-hydroxysteroid
- 7-hydroxysteroid
- 7-alpha-hydroxysteroid
- 3-alpha-hydroxysteroid
- Hydroxysteroid
- Cyclic alcohol
- Secondary alcohol
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxide
- Alcohol
- Organic oxygen compound
- Carbonyl group
- Hydrocarbon derivative
- Organooxygen compound
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-01rt-0419000000-6a92f910581240163a99 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-0006-1110390000-52bc66ab11fd14425d4f | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_3_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-qTof , Positive | splash10-0a4l-3940000000-cf90a6f216d592e2ad4c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , negative | splash10-00di-0029000000-54929e08fef761ba2b28 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - , positive | splash10-0002-2911000000-eec2b269eeb08a30ac6e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-056r-0009000000-997e61e986e67265241c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-056r-0009000000-81134947694f847d1c65 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-02t9-1219000000-6a0ddebebacac090c27e | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0009000000-efdaad69eee0ae934dd5 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-006x-1009000000-15b4edab9d05e4c3693a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9006000000-9291e69db3a3c47ecd97 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-0009000000-7631731446db77938069 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-006x-0009000000-39e7e555dc6b36ada2af | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000i-1009000000-351c21e3dab693818b22 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0019000000-56dc67f71a8507c1433f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-3039000000-510702cae19e3906b6e1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0002-9550000000-7486a5f56afd302efa7e | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, CDCl3, experimental) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, CD3OD, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|