| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:31:09 UTC |
|---|
| Update Date | 2020-05-11 20:20:57 UTC |
|---|
| BMDB ID | BMDB0000467 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Nutriacholic acid |
|---|
| Description | Nutriacholic acid, also known as nutriacholate, belongs to the class of organic compounds known as monohydroxy bile acids, alcohols and derivatives. These are bile acids, alcohols or any of their derivatives bearing a hydroxyl group. Based on a literature review a significant number of articles have been published on Nutriacholic acid. |
|---|
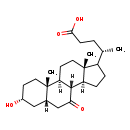
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Nutriacholate | Generator | | 3a-Hydroxy-7-oxo-5b-cholanoate | HMDB | | 3a-Hydroxy-7-oxo-5b-cholanoic acid | HMDB | | 5b-Cholanic acid-3a-ol-7-one | HMDB | | 7-Ketochenodeoxycholate | HMDB | | 7-Ketochenodeoxycholic acid | HMDB | | 7-Ketolithocholate | HMDB | | 7-Ketolithocholic acid | HMDB, MeSH | | 7-oxo-3a-Hydroxycholan-24-Oate | HMDB | | 7-oxo-3a-Hydroxycholan-24-Oic acid | HMDB | | 7-Oxolithocholate | HMDB | | 7-Oxolithocholic acid | HMDB, MeSH | | 3 alpha-Hydroxy-7-keto-5 beta-cholanoate | MeSH, HMDB | | 3 alpha-Ol-7-one-5 beta-cholanoic acid | MeSH, HMDB | | 7-Ketolithocholic acid, (3beta,5alpha)-isomer | MeSH, HMDB | | (4S)-4-[(1S,2S,5R,7S,10R,11S,15R)-5-Hydroxy-2,15-dimethyl-9-oxotetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]pentanoate | Generator, HMDB |
|
|---|
| Chemical Formula | C24H38O4 |
|---|
| Average Molecular Weight | 390.5561 |
|---|
| Monoisotopic Molecular Weight | 390.277009704 |
|---|
| IUPAC Name | (4S)-4-[(1S,2S,5R,7S,10R,11S,15R)-5-hydroxy-2,15-dimethyl-9-oxotetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadecan-14-yl]pentanoic acid |
|---|
| Traditional Name | (4S)-4-[(1S,2S,5R,7S,10R,11S,15R)-5-hydroxy-2,15-dimethyl-9-oxotetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadecan-14-yl]pentanoic acid |
|---|
| CAS Registry Number | 4651-67-6 |
|---|
| SMILES | [H][C@@]12CCC([C@@H](C)CCC(O)=O)[C@@]1(C)CC[C@@]1([H])[C@@]2([H])C(=O)C[C@]2([H])C[C@H](O)CC[C@]12C |
|---|
| InChI Identifier | InChI=1S/C24H38O4/c1-14(4-7-21(27)28)17-5-6-18-22-19(9-11-24(17,18)3)23(2)10-8-16(25)12-15(23)13-20(22)26/h14-19,22,25H,4-13H2,1-3H3,(H,27,28)/t14-,15-,16+,17?,18-,19-,22-,23-,24+/m0/s1 |
|---|
| InChI Key | DXOCDBGWDZAYRQ-QPVZPPSOSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as monohydroxy bile acids, alcohols and derivatives. These are bile acids, alcohols or any of their derivatives bearing a hydroxyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Bile acids, alcohols and derivatives |
|---|
| Direct Parent | Monohydroxy bile acids, alcohols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Monohydroxy bile acid, alcohol, or derivatives
- 3-hydroxysteroid
- Hydroxysteroid
- 7-oxosteroid
- 3-alpha-hydroxysteroid
- Oxosteroid
- Cyclic alcohol
- Secondary alcohol
- Ketone
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxide
- Alcohol
- Organic oxygen compound
- Carbonyl group
- Hydrocarbon derivative
- Organooxygen compound
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03gj-0319000000-99c0cf456750a298fdba | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-01b9-2141590000-df782bc25f3fffc7b8b0 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-014i-0291000000-9f5bf5f94dd6944d6d6f | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-052r-2962000000-f18d1686d1ac8b6e93f3 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-05o1-2900000000-681b2fd8ee33b91d7a44 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-05fr-0009000000-96e2b732bac67ff31b5c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-076s-0009000000-63349742d082a77d840f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01t9-0229000000-c96c7b6b3568bc1568ec | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0009000000-bcd0337fb08276e5242b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00dr-0009000000-8e05a17c162e99bd54ed | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9006000000-ddfed24aad7a991420fa | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0009000000-1bf3e4a9164eecd8b5ad | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0079-0009000000-dabdf5ac37340fa8a343 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052r-3029000000-ec70dabf7a7d2522ff70 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0596-0009000000-aa05b3718c19db13d245 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0ab9-2059000000-993d476f495649000a3f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0adj-2931000000-238650ed3f34647466eb | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, CDCl3, experimental) | Not Available | View in JSpectraViewer |
|---|
| 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, CD3OD, experimental) | Not Available | View in JSpectraViewer |
|---|
|
|---|