| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:30:13 UTC |
|---|
| Update Date | 2020-04-22 15:03:27 UTC |
|---|
| BMDB ID | BMDB0000409 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
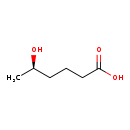
| Common Name | (5R)-5-Hydroxyhexanoic acid |
|---|
| Description | (5R)-5-Hydroxyhexanoic acid, also known as (5R)-5-hydroxycaproate or HMBA, belongs to the class of organic compounds known as medium-chain fatty acids. These are fatty acids with an aliphatic tail that contains between 4 and 12 carbon atoms. Based on a literature review very few articles have been published on (5R)-5-Hydroxyhexanoic acid. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (-)-5-Hydroxycaproic acid | ChEBI | | (-)-5-Hydroxyhexanoic acid | ChEBI | | (5R)-5-Hydroxycaproic acid | ChEBI | | (R)-(-)-5-Hydroxyhexanoic acid | ChEBI | | (-)-5-Hydroxycaproate | Generator | | (-)-5-Hydroxyhexanoate | Generator | | (5R)-5-Hydroxycaproate | Generator | | (R)-(-)-5-Hydroxyhexanoate | Generator | | (5R)-5-Hydroxyhexanoate | Generator | | (5R)-5-Hydroxy-hexanoate | HMDB | | (5R)-5-Hydroxy-hexanoic acid | HMDB | | HMBA | HMDB | | 5R-Hydroxy-hexanoate | HMDB |
|
|---|
| Chemical Formula | C6H12O3 |
|---|
| Average Molecular Weight | 132.1577 |
|---|
| Monoisotopic Molecular Weight | 132.07864425 |
|---|
| IUPAC Name | (5R)-5-hydroxyhexanoic acid |
|---|
| Traditional Name | HMBA |
|---|
| CAS Registry Number | 83972-61-6 |
|---|
| SMILES | C[C@@H](O)CCCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H12O3/c1-5(7)3-2-4-6(8)9/h5,7H,2-4H2,1H3,(H,8,9)/t5-/m1/s1 |
|---|
| InChI Key | YDCRNMJQROAWFT-RXMQYKEDSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as medium-chain fatty acids. These are fatty acids with an aliphatic tail that contains between 4 and 12 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acids and conjugates |
|---|
| Direct Parent | Medium-chain fatty acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Medium-chain fatty acid
- Hydroxy fatty acid
- Secondary alcohol
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Adiposome
- Cell membrane
- Cytoplasm
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0077-9100000000-13a584fc9ff05431f359 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (2 TMS) - 70eV, Positive | splash10-007c-9420000000-7b0146069089183f6082 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-5900000000-3bf905a3ced857b422b1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014i-9400000000-b18891bb2981129af001 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-066r-9000000000-3510777085e9e83a6561 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-2900000000-3b9f578bc70674738913 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03e9-4900000000-9944af827f01e265b301 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-9f3f80d6749af1cc6dbb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03ei-3900000000-49a5b195fc6d1869f127 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0bta-9400000000-9da83e480e59bbc1ade7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052g-9000000000-e9748f05fe518d153135 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-9400000000-744ecd7d7e40b1357594 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05tk-9000000000-086484b097d1ba55d5d0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052f-9000000000-db58a877cf5945d523f9 | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|
| Synthesis Reference | Fazio, Fabio; Schneider, Manfred P. FA-catalyzed trimerization of R-(+)-6-methyltetrahydro-2-pyranone. FB 9-Bergische Universitat-GH-Wuppertal, Wuppertal, Germany. Tetrahedron Letters (2002), 43(5), 811-814. |
|---|