| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:30:06 UTC |
|---|
| Update Date | 2020-04-22 15:03:24 UTC |
|---|
| BMDB ID | BMDB0000403 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 2-Hydroxyadenine |
|---|
| Description | 2-Hydroxyadenine, also known as 2-OH-ade or isoguanine, belongs to the class of organic compounds known as 6-aminopurines. These are purines that carry an amino group at position 6. Purine is a bicyclic aromatic compound made up of a pyrimidine ring fused to an imidazole ring. Based on a literature review a significant number of articles have been published on 2-Hydroxyadenine. |
|---|
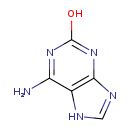
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-Oxoadenine | ChEBI | | 6-Amino-1,3-dihydro-2H-purin-2-one | ChEBI | | 6-Amino-3,7-dihydro-purin-2-one | ChEBI | | 6-Amino-3,9-dihydro-2H-purin-2-one | ChEBI | | 6-Amino-7H-purin-2-ol | ChEBI | | 6-Amino-9H-purin-2-ol | ChEBI | | 2-Hydroxy-6-aminopurine | HMDB | | 2-Oxyadenine | HMDB | | 6-Amino-2-hydroxypurine | HMDB | | 6-Amino-3,7(9)-dihydro-purin-2-one | HMDB | | Isoguanine | HMDB | | 2-OH-Ade | HMDB | | Crotonoside | HMDB | | 2-Hydroxyadenine | ChEBI |
|
|---|
| Chemical Formula | C5H5N5O |
|---|
| Average Molecular Weight | 151.1261 |
|---|
| Monoisotopic Molecular Weight | 151.049409807 |
|---|
| IUPAC Name | 6-amino-7H-purin-2-ol |
|---|
| Traditional Name | 2-hydroxy-6-aminopurine |
|---|
| CAS Registry Number | 3373-53-3 |
|---|
| SMILES | NC1=NC(O)=NC2=C1NC=N2 |
|---|
| InChI Identifier | InChI=1S/C5H5N5O/c6-3-2-4(8-1-7-2)10-5(11)9-3/h1H,(H4,6,7,8,9,10,11) |
|---|
| InChI Key | DRAVOWXCEBXPTN-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as 6-aminopurines. These are purines that carry an amino group at position 6. Purine is a bicyclic aromatic compound made up of a pyrimidine ring fused to an imidazole ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | 6-aminopurines |
|---|
| Alternative Parents | |
|---|
| Substituents | - 6-aminopurine
- Aminopyrimidine
- Hydroxypyrimidine
- Pyrimidine
- Imidolactam
- Azole
- Imidazole
- Heteroaromatic compound
- Azacycle
- Organopnictogen compound
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Amine
- Organic nitrogen compound
- Hydrocarbon derivative
- Organic oxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | > 360 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 0.0625 mg/mL | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0udi-1900000000-c16950878c62efe1b98d | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-0kmi-7930000000-f60855003c167d5a6142 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0udi-0900000000-42af2345f291c0931747 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-053r-2900000000-20dc64d93a8160bc7cba | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0kai-9200000000-dd4e5a7f42bbde9a7362 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900000000-20dd242c9f0be41ca3b9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0900000000-0a5cc033a455487be201 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a59-9700000000-88fd5f0f4ed191155fca | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0900000000-db76d46c9a9e58708daf | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0pb9-1900000000-2c2e928303dd65e7b74a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001l-9100000000-cba052229db1704e3934 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900000000-361a2474138028086bb3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0900000000-c0348939b06d3f4c02ac | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9400000000-23cc92eaf9fad62612b8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0900000000-21c3d82f91283a8a49e6 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0udi-2900000000-abe3ac9c0f2bdc1aff6c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9100000000-3ba35f09b305bcf5a6cd | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|