Showing metabocard for 2,8-Dihydroxyadenine (BMDB0000401)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Creation Date | 2016-09-30 22:30:03 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 2020-04-22 15:03:24 UTC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BMDB ID | BMDB0000401 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Secondary Accession Numbers |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Common Name | 2,8-Dihydroxyadenine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | 2,8-Dihydroxyadenine, also known as 6-amino-purine-2,8-diol or dimethyloctanoate, belongs to the class of organic compounds known as purinones. These are purines in which the purine moiety bears a C=O group. Purine is a bicyclic aromatic compound made up of a pyrimidine ring fused to an imidazole ring. Based on a literature review a significant number of articles have been published on 2,8-Dihydroxyadenine. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
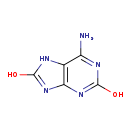
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula | C5H5N5O2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight | 167.1255 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight | 167.044324429 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name | 6-amino-7H-purine-2,8-diol | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional Name | 6-amino-2,8-dihydroxypurine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS Registry Number | 30377-37-8 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES | NC1=NC(O)=NC2=C1NC(O)=N2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Identifier | InChI=1S/C5H5N5O2/c6-2-1-3(9-4(11)7-1)10-5(12)8-2/h(H5,6,7,8,9,10,11,12) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key | XFBOJHLYDJZYSP-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description | belongs to the class of organic compounds known as purinones. These are purines in which the purine moiety bears a C=O group. Purine is a bicyclic aromatic compound made up of a pyrimidine ring fused to an imidazole ring. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organoheterocyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Imidazopyrimidines | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Purines and purine derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Purinones | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Ontology | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Status | Expected but not Quantified | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Origin |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biofunction | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Application | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State | Solid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biospecimen Locations | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Normal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Abnormal Concentrations | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| HMDB ID | HMDB0000401 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DrugBank ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Phenol Explorer Compound ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| FooDB ID | FDB022018 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KNApSAcK ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemspider ID | 83302 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Compound ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BioCyc ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| BiGG ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikipedia Link | 2,8-Dihydroxyadenine | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| METLIN ID | 5390 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PubChem Compound | 92268 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PDB ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ChEBI ID | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference | Stevens, Marcus A.; Smith, Herman W.; Brown, George Bosworth. Purine N-oxides. VII. Reaction of aminopurine 1-N-oxides with acetic anhydride. Journal of the American Chemical Society (1960), 82 1148-52. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General References | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||