| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:30:02 UTC |
|---|
| Update Date | 2020-05-11 20:20:45 UTC |
|---|
| BMDB ID | BMDB0000400 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 3,7-Dihydroxy-12-oxocholanoic acid |
|---|
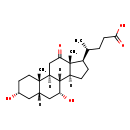
| Description | 3,7-Dihydroxy-12-oxocholanoic acid, also known as 3α,7α-dihydroxy-12-oxo-5β-cholanate or 12-keto-chenodeoxycholate, belongs to the class of organic compounds known as dihydroxy bile acids, alcohols and derivatives. Dihydroxy bile acids, alcohols and derivatives are compounds containing or derived from a bile acid or alcohol, and which bears exactly two carboxylic acid groups. 3,7-Dihydroxy-12-oxocholanoic acid is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (3alpha,5beta,7alpha)-3,7-Dihydroxy-12-oxocholan-24-Oic acid | ChEBI | | 3alpha,7alpha-Dihydroxy-12-oxo-5beta-cholanate | ChEBI | | (3a,5b,7a)-3,7-Dihydroxy-12-oxocholan-24-Oate | Generator | | (3a,5b,7a)-3,7-Dihydroxy-12-oxocholan-24-Oic acid | Generator | | (3alpha,5beta,7alpha)-3,7-Dihydroxy-12-oxocholan-24-Oate | Generator | | (3Α,5β,7α)-3,7-dihydroxy-12-oxocholan-24-Oate | Generator | | (3Α,5β,7α)-3,7-dihydroxy-12-oxocholan-24-Oic acid | Generator | | 3a,7a-Dihydroxy-12-oxo-5b-cholanate | Generator | | 3a,7a-Dihydroxy-12-oxo-5b-cholanic acid | Generator | | 3alpha,7alpha-Dihydroxy-12-oxo-5beta-cholanic acid | Generator | | 3Α,7α-dihydroxy-12-oxo-5β-cholanate | Generator | | 3Α,7α-dihydroxy-12-oxo-5β-cholanic acid | Generator | | 3,7-Dihydroxy-12-oxocholanoate | Generator | | (3a,5b,7a)-3,7-Dihydroxy-12-oxo-cholan-24-Oate | HMDB | | (3a,5b,7a)-3,7-Dihydroxy-12-oxo-cholan-24-Oic acid | HMDB | | 12-Keto-3a,7a-dihydroxy-5b-cholanate | HMDB | | 12-Keto-3a,7a-dihydroxy-5b-cholanic acid | HMDB | | 12-Keto-chenodeoxycholate | HMDB | | 12-Keto-chenodeoxycholic acid | HMDB | | 12-Ketochenodeoxycholate | HMDB | | 12-Ketochenodeoxycholic acid | HMDB | | 12-oxo-3a,7a-Dihydroxy-5b-cholan-24-Oate | HMDB | | 12-oxo-3a,7a-Dihydroxy-5b-cholan-24-Oic acid | HMDB | | 12-Oxochenodeoxycholate | HMDB | | 12-Oxochenodeoxycholic acid | HMDB | | 3a,7a-Dihydroxy-12-keto-5b-cholanate | HMDB | | 3a,7a-Dihydroxy-12-keto-5b-cholanic acid | HMDB | | 3a,7a-Dihydroxy-12-ketocholanate | HMDB | | 3a,7a-Dihydroxy-12-ketocholanic acid | HMDB | | 3a,7a-Dihydroxy-12-ketocholate | HMDB | | 3a,7a-Dihydroxy-12-ketocholic acid | HMDB | | 3a,7a-Dihydroxy-12-oxo-5b-cholan-24-Oate | HMDB | | 3a,7a-Dihydroxy-12-oxo-5b-cholan-24-Oic acid | HMDB | | 3a,7a-Dihydroxy-12-oxo-5b-cholanoate | HMDB | | 3a,7a-Dihydroxy-12-oxo-5b-cholanoic acid | HMDB | | 3a,7a-Dihydroxy-12-oxocholate | HMDB | | 3a,7a-Dihydroxy-12-oxocholic acid | HMDB | | 5b-Cholanic acid-3a,7a-diol-12-one | HMDB | | 12-Monoketocholate | HMDB | | 12-Monoketocholic acid | HMDB | | 12-oxo-3,7-Dihydroxycholanoic acid | HMDB | | 3 alpha, 7 alpha-Dihydroxy-12-one-5 beta-cholanoic acid | HMDB | | 3 alpha,7 alpha-Dihydroxy-12-keto-5 beta-cholanoic acid | HMDB | | 3alpha,7alpha-Dihydroxy-12-keto-5beta-cholanoic acid | HMDB | | Sodium monoketocholate | HMDB | | (4R)-4-[(1S,2S,5R,7S,9R,10R,11S,14R,15R)-5,9-Dihydroxy-2,15-dimethyl-16-oxotetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]pentanoate | HMDB | | 3,7-Dihydroxy-12-oxocholanoic acid | ChEBI |
|
|---|
| Chemical Formula | C24H38O5 |
|---|
| Average Molecular Weight | 406.5555 |
|---|
| Monoisotopic Molecular Weight | 406.271924326 |
|---|
| IUPAC Name | (4R)-4-[(1S,2S,5R,7S,9R,10R,11S,14R,15R)-5,9-dihydroxy-2,15-dimethyl-16-oxotetracyclo[8.7.0.0²,⁷.0¹¹,¹⁵]heptadecan-14-yl]pentanoic acid |
|---|
| Traditional Name | 12-ketochenodeoxycholic acid |
|---|
| CAS Registry Number | 2458-08-4 |
|---|
| SMILES | [H][C@@]12CC[C@H]([C@H](C)CCC(O)=O)[C@@]1(C)C(=O)C[C@@]1([H])[C@@]2([H])[C@H](O)C[C@]2([H])C[C@H](O)CC[C@]12C |
|---|
| InChI Identifier | InChI=1S/C24H38O5/c1-13(4-7-21(28)29)16-5-6-17-22-18(12-20(27)24(16,17)3)23(2)9-8-15(25)10-14(23)11-19(22)26/h13-19,22,25-26H,4-12H2,1-3H3,(H,28,29)/t13-,14+,15-,16-,17+,18+,19-,22+,23+,24-/m1/s1 |
|---|
| InChI Key | MIHNUBCEFJLAGN-DMMBONCOSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as dihydroxy bile acids, alcohols and derivatives. Dihydroxy bile acids, alcohols and derivatives are compounds containing or derived from a bile acid or alcohol, and which bears exactly two carboxylic acid groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Bile acids, alcohols and derivatives |
|---|
| Direct Parent | Dihydroxy bile acids, alcohols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Dihydroxy bile acid, alcohol, or derivatives
- 3-hydroxysteroid
- 7-hydroxysteroid
- 3-alpha-hydroxysteroid
- Oxosteroid
- 12-oxosteroid
- Hydroxysteroid
- Cyclic alcohol
- Secondary alcohol
- Ketone
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxide
- Alcohol
- Organic oxygen compound
- Carbonyl group
- Hydrocarbon derivative
- Organooxygen compound
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-03e9-0629000000-3135db41ff8b968182b8 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (3 TMS) - 70eV, Positive | splash10-0a4i-2201198000-bb38056a165602f5feb7 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_2_6) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TMS_3_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_1_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_1) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_2) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_3) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_4) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (TBDMS_2_5) - 70eV, Positive | Not Available | View in JSpectraViewer |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0079-0009100000-bccc650b1cd4508f1aa7 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-00dr-0009000000-c0c7d7baa207e3cc0a05 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-056u-1109000000-e710af896d258d339fef | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4r-0008900000-dd8b045786455a7b2b5f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-052r-1009300000-40bda8fb29d7dc21b644 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9006000000-879f42792050e0f3d07a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0002900000-042ac9446c06cee5ac0f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4r-0009700000-eec83a5cd15c56ccdff9 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0udi-1039100000-e4f72f94d4744ca5a96c | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-059i-0009300000-f1bc8868b74d47d136be | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0079-1169100000-6bf02d9105b1f9389f99 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9561000000-ee15985d116087cd50ec | View in MoNA |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|