| Record Information |
|---|
| Version | 1.0 |
|---|
| Creation Date | 2016-09-30 22:28:07 UTC |
|---|
| Update Date | 2020-05-11 20:50:29 UTC |
|---|
| BMDB ID | BMDB0000304 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | Uridine diphosphate-N-acetylgalactosamine |
|---|
| Description | Uridine diphosphate-N-acetylgalactosamine belongs to the class of organic compounds known as pyrimidine nucleotide sugars. These are pyrimidine nucleotides bound to a saccharide derivative through the terminal phosphate group. Based on a literature review very few articles have been published on Uridine diphosphate-N-acetylgalactosamine. |
|---|
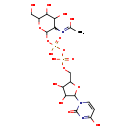
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Uridine diphosphoric acid-N-acetylgalactosamine | Generator | | UDP-N-Acetyl-D-galactosamine | HMDB | | UDP-N-Acetyl-delta-galactosamine | HMDB | | UDP-N-Acetylgalactosamine | HMDB | | Uridine 5'-diphospho-N-acetylgalactosamine | HMDB | | Uridine diphosphate-N-acetyl-D-galactosamine | HMDB | | Uridine diphosphate-N-acetyl-delta-galactosamine | HMDB | | Uridine diphospho-2-acetamido-2-deoxy-D-galactose | HMDB | | Uridine diphospho-2-acetamido-2-deoxy-delta-galactose | HMDB | | Uridine diphospho-N-acetylgalactosamine | HMDB | | Uridine diphosphoacetylgalactosamine | HMDB | | Uridine pyrophosphate 2-acetamido-2-deoxy-a-D-galactopyranosyl ester | HMDB | | Uridine pyrophosphate 2-acetamido-2-deoxy-alpha-D-galactopyranosyl ester | HMDB | | Uridine pyrophosphate 2-acetamido-2-deoxy-alpha-delta-galactopyranosyl ester | HMDB | | Uridine pyrophosphate N-acetyl-a-D-chondrosamine ester | HMDB | | Uridine pyrophosphate N-acetyl-alpha-D-chondrosamine ester | HMDB | | Uridine pyrophosphate N-acetyl-alpha-delta-chondrosamine ester | HMDB | | Uridine pyrophosphoacetylgalactosamine | HMDB | | Pyrophosphoacetylglucosamine, uridine | MeSH, HMDB | | UDP Acetylglucosamine | MeSH, HMDB | | Uridine diphospho-N-acetylglucosamine | MeSH, HMDB | | Acetylgalactosamine, UDP | MeSH, HMDB | | diphospho-N-Acetylglucosamine, uridine | MeSH, HMDB | | Uridine diphosphate N acetylgalactosamine | MeSH, HMDB | | Uridine diphosphate N acetylglucosamine | MeSH, HMDB | | Uridine diphosphate N-acetylglucosamine | MeSH, HMDB | | Diphosphate N-acetylglucosamine, uridine | MeSH, HMDB | | UDPGNAc | MeSH, HMDB | | Uridine diphosphate N-acetylgalactosamine | MeSH, HMDB | | Acetylglucosamine, UDP | MeSH, HMDB | | N-Acetylgalactosamine, uridine diphosphate | MeSH, HMDB | | UDP Acetylgalactosamine | MeSH, HMDB | | Uridine pyrophosphoacetylglucosamine | MeSH, HMDB | | Uridine diphospho N acetylglucosamine | MeSH, HMDB | | Diphosphate N-acetylgalactosamine, uridine | MeSH, HMDB | | N-Acetylglucosamine, uridine diphosphate | MeSH, HMDB | | N-[2-({[({[3,4-dihydroxy-5-(4-hydroxy-2-oxo-1,2-dihydropyrimidin-1-yl)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)-4,5-dihydroxy-6-(hydroxymethyl)oxan-3-yl]ethanimidate | Generator, HMDB |
|
|---|
| Chemical Formula | C17H27N3O17P2 |
|---|
| Average Molecular Weight | 607.3537 |
|---|
| Monoisotopic Molecular Weight | 607.081569477 |
|---|
| IUPAC Name | [({[5-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]({[3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy})phosphinic acid |
|---|
| Traditional Name | {[5-(2,4-dioxo-3H-pyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy([3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy)phosphinic acid |
|---|
| CAS Registry Number | 7277-98-7 |
|---|
| SMILES | CC(O)=NC1C(OP(O)(=O)OP(O)(=O)OCC2OC(C(O)C2O)N2C=CC(O)=NC2=O)OC(CO)C(O)C1O |
|---|
| InChI Identifier | InChI=1S/C17H27N3O17P2/c1-6(22)18-10-13(26)11(24)7(4-21)35-16(10)36-39(31,32)37-38(29,30)33-5-8-12(25)14(27)15(34-8)20-3-2-9(23)19-17(20)28/h2-3,7-8,10-16,21,24-27H,4-5H2,1H3,(H,18,22)(H,29,30)(H,31,32)(H,19,23,28) |
|---|
| InChI Key | LFTYTUAZOPRMMI-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | belongs to the class of organic compounds known as pyrimidine nucleotide sugars. These are pyrimidine nucleotides bound to a saccharide derivative through the terminal phosphate group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Pyrimidine nucleotides |
|---|
| Sub Class | Pyrimidine nucleotide sugars |
|---|
| Direct Parent | Pyrimidine nucleotide sugars |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine nucleotide sugar
- Pyrimidine ribonucleoside diphosphate
- Pentose phosphate
- Pentose-5-phosphate
- N-acyl-alpha-hexosamine
- Glycosyl compound
- N-glycosyl compound
- Monosaccharide phosphate
- Organic pyrophosphate
- Pyrimidone
- Monoalkyl phosphate
- Hydropyrimidine
- Monosaccharide
- Organic phosphoric acid derivative
- Oxane
- Phosphoric acid ester
- Pyrimidine
- Alkyl phosphate
- Vinylogous amide
- Acetamide
- Tetrahydrofuran
- Heteroaromatic compound
- Urea
- Secondary carboxylic acid amide
- Secondary alcohol
- Carboxamide group
- Lactam
- Azacycle
- Carboxylic acid derivative
- Oxacycle
- Organoheterocyclic compound
- Organopnictogen compound
- Organooxygen compound
- Alcohol
- Primary alcohol
- Organic nitrogen compound
- Organic oxygen compound
- Organic oxide
- Carbonyl group
- Hydrocarbon derivative
- Organonitrogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | Not Available |
|---|
| Ontology |
|---|
| Status | Expected but not Quantified |
|---|
| Origin | |
|---|
| Biofunction | Not Available |
|---|
| Application | Not Available |
|---|
| Cellular locations | - Endoplasmic reticulum
- Golgi
- Lysosome
|
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties | |
|---|
| Spectra |
|---|
| Spectra | | Spectrum Type | Description | Splash Key | |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (Non-derivatized) - 70eV, Positive | splash10-0f96-5924660000-c11cc7fa21a050c0f373 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS (1 TMS) - 70eV, Positive | splash10-03fu-5926205000-cdab8d90ff64fb776a15 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - , negative | splash10-05e9-8897100000-095b6091bd84d37fd58b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - , positive | splash10-000l-2900000000-82a9c4130de4b3125fd7 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-000l-2900000000-4d24529e74ad2fa21c05 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0f79-1930010000-55436553b484e5220f19 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-1490000000-ac1c456c4492cae878a2 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0910010000-188f1b92ae056d7c7cd0 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-3921000000-ce6262fadd328364b376 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-5910000000-b91dd47ef36e2cad815b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-06ri-9502321000-f502248d5c26ca57aecb | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01ox-9605010000-08fa474eedca91ebd68f | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-06tf-5911000000-55e7efddb090fc85714a | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0bt9-0000095000-b04755a1d58b54039931 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-08fr-4201191000-b741fdd379d9a2afcc61 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-053u-5719010000-abc6f96328a6d0fbfbb8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-056r-0000914000-04cfea1597e2554c0ff8 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-056r-1412944000-aafe333f4e24af5bf092 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0h3r-9231000000-e1c43f13a820bf095dda | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | Not Available | View in JSpectraViewer |
|---|
|
|---|